11.3 Utilizing two predictors
What we know about afternoon temperatures in Australia:
- positively associated with morning temperatures
- tend to be lower in Wollongong than in Uluru
Now extend the model to:
two-predictor model of 3 p.m. temperature using both temp9am and location
ggplot(weather_WU, aes(y = temp3pm,
x = temp9am,
color = location)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE)## `geom_smooth()` using formula = 'y ~ x'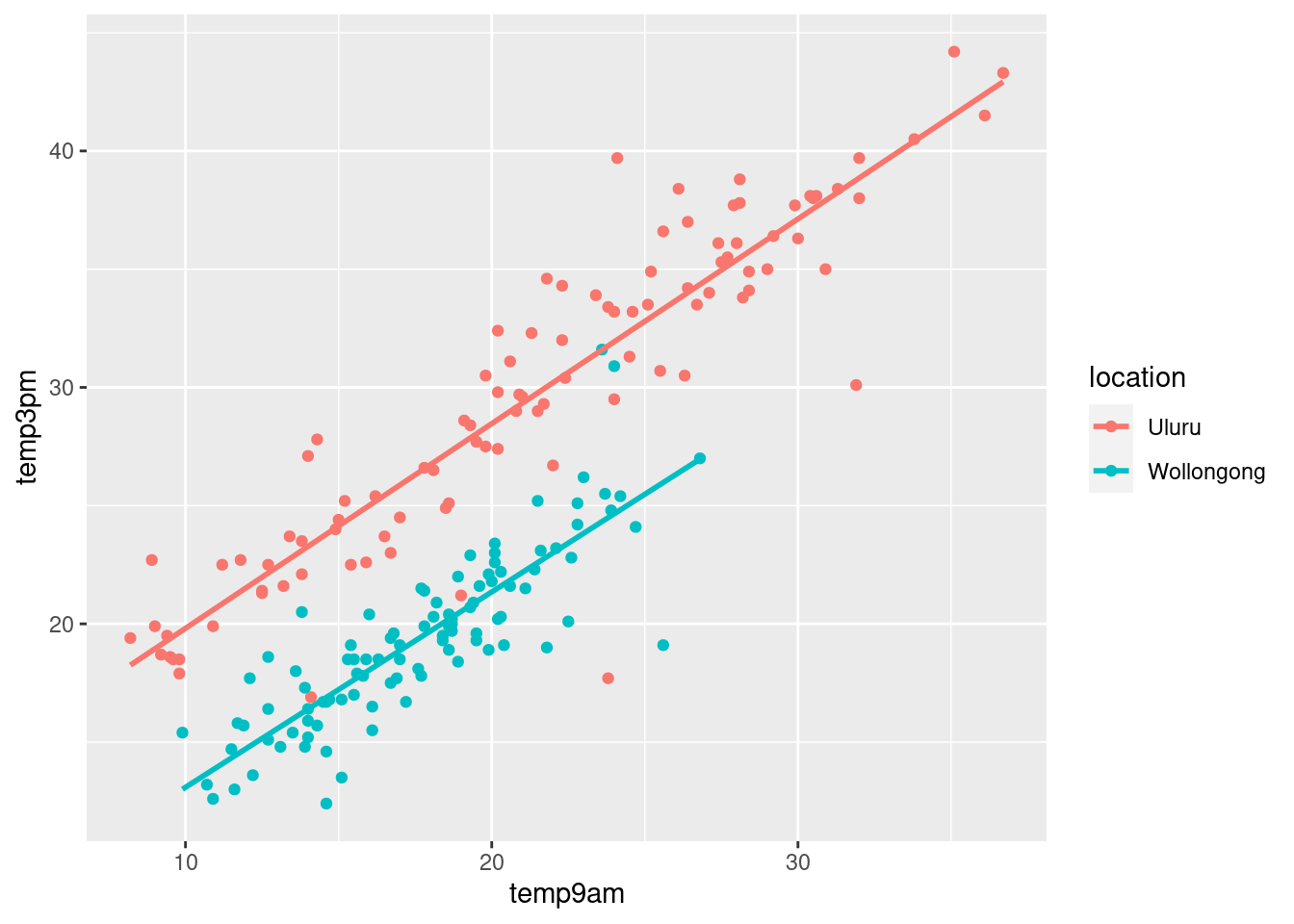
weather_model_3_prior <- stan_glm(
temp3pm ~ temp9am + location,
data = weather_WU,
family = gaussian,
prior_intercept = normal(25, 5),
prior = normal(0, 2.5, autoscale = TRUE),
prior_aux = exponential(1, autoscale = TRUE),
chains = 4,
iter = 5000*2,
seed = 84735,
prior_PD = TRUE)11.3.1 Simulate 100 datasets from the prior models
set.seed(84735)
weather_WU %>%
add_predicted_draws(weather_model_3_prior, n = 100) %>%
ggplot(aes(x = .prediction, group = .draw)) +
geom_density() +
xlab("temp3pm")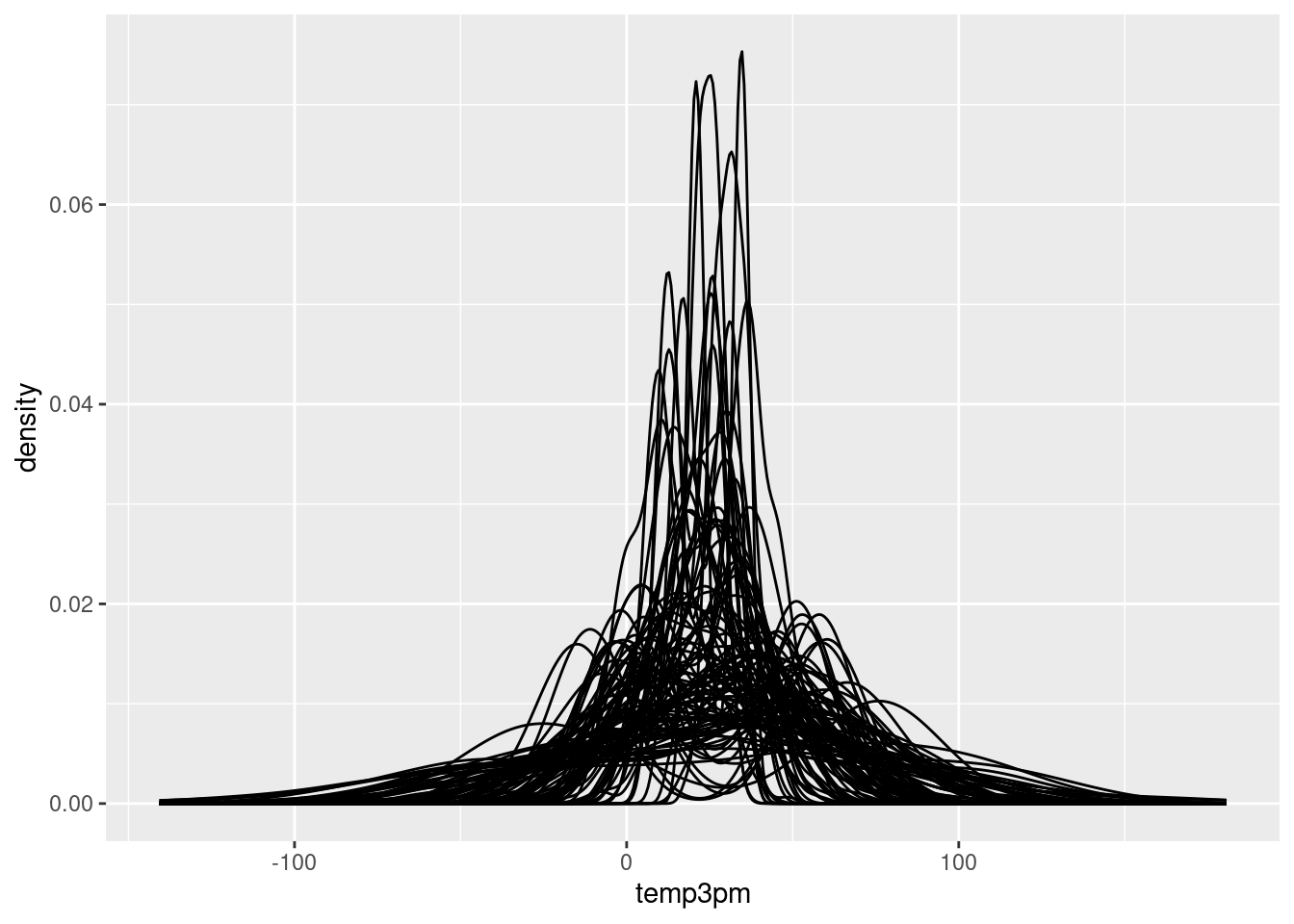
weather_WU %>%
add_fitted_draws(weather_model_3_prior, n = 100) %>%
ggplot(aes(x = temp9am,
y = temp3pm,
color = location)) +
geom_line(aes(y = .value,
group = paste(location, .draw)))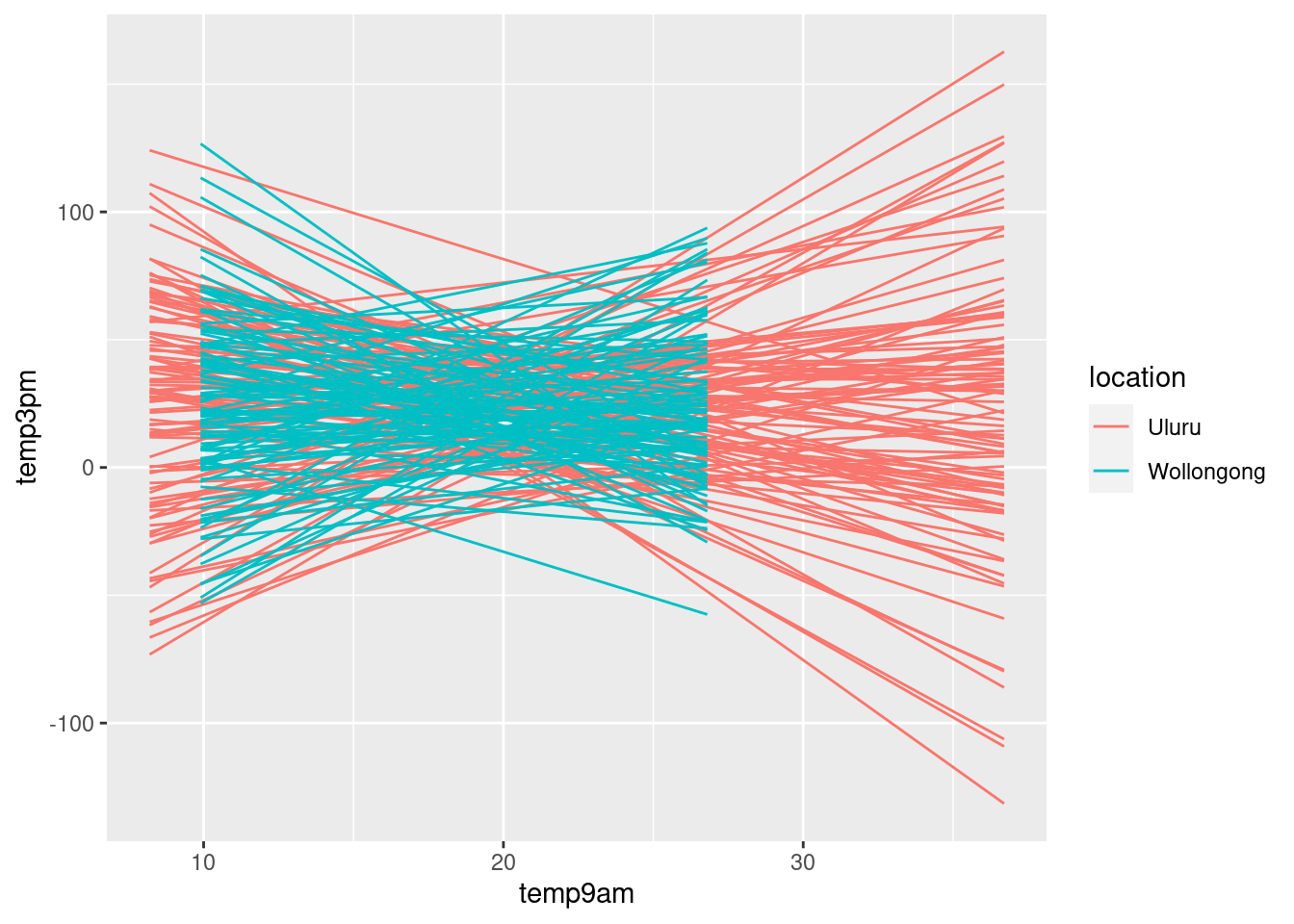
Combine prior assumption with simulations, specifying
prior_PD = FALSEweather_model_3 <- update(weather_model_3_prior,
prior_PD = FALSE)Across all 100 scenarios, 3 p.m. temperature is positively associated with 9 a.m. temperature and tends to be higher in Uluru than in Wollongong.
head(as.data.frame(weather_model_3), 6)## (Intercept) temp9am locationWollongong sigma
## 1 13.04657 0.8016891 -7.663020 2.392109
## 2 12.73136 0.8174191 -7.838690 2.444850
## 3 11.81295 0.8615380 -7.647937 2.413606
## 4 12.37991 0.8041587 -6.845878 2.552823
## 5 12.22305 0.8391698 -7.419569 2.498952
## 6 10.94792 0.8539339 -6.821811 2.272895weather_WU %>%
add_fitted_draws(weather_model_3, n = 100) %>%
ggplot(aes(x = temp9am, y = temp3pm, color = location)) +
geom_line(aes(y = .value, group = paste(location, .draw)), alpha = .1) +
geom_point(data = weather_WU, size = 0.1)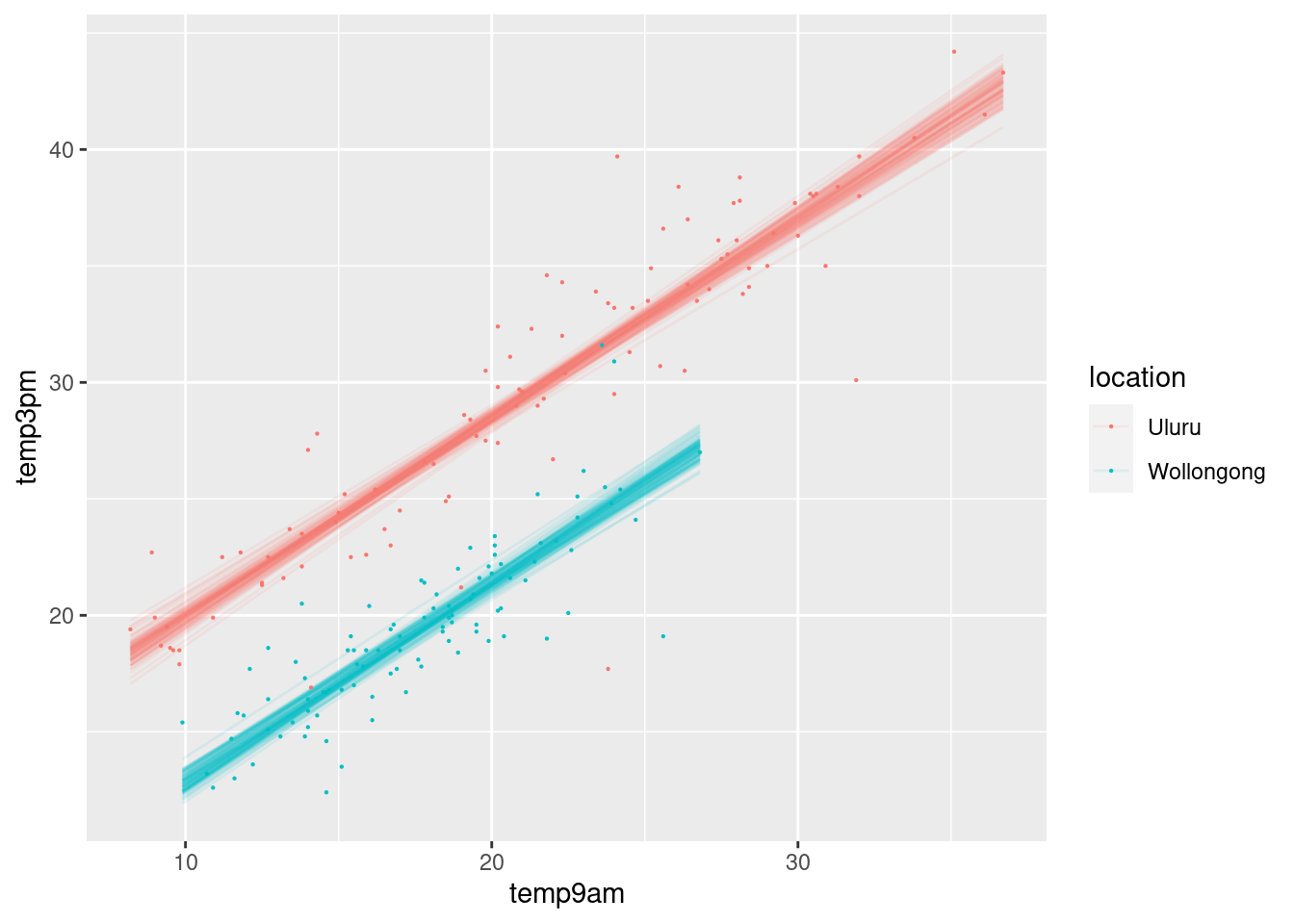
# Posterior summaries
posterior_interval(weather_model_3, prob = 0.80,
pars = c("temp9am", "locationWollongong"))## 10% 90%
## temp9am 0.8196281 0.8945346
## locationWollongong -7.5067716 -6.599905711.3.2 Predict 3 p.m. temperature on specific days
# Simulate a set of predictions
set.seed(84735)
temp3pm_prediction <- posterior_predict(
weather_model_3,
newdata = data.frame(temp9am = c(10, 10),
location = c("Uluru", "Wollongong")))# Plot the posterior predictive models
mcmc_areas(temp3pm_prediction) +
ggplot2::scale_y_discrete(labels = c("Uluru", "Wollongong")) +
xlab("temp3pm")## Scale for y is already present.
## Adding another scale for y, which will replace the existing scale.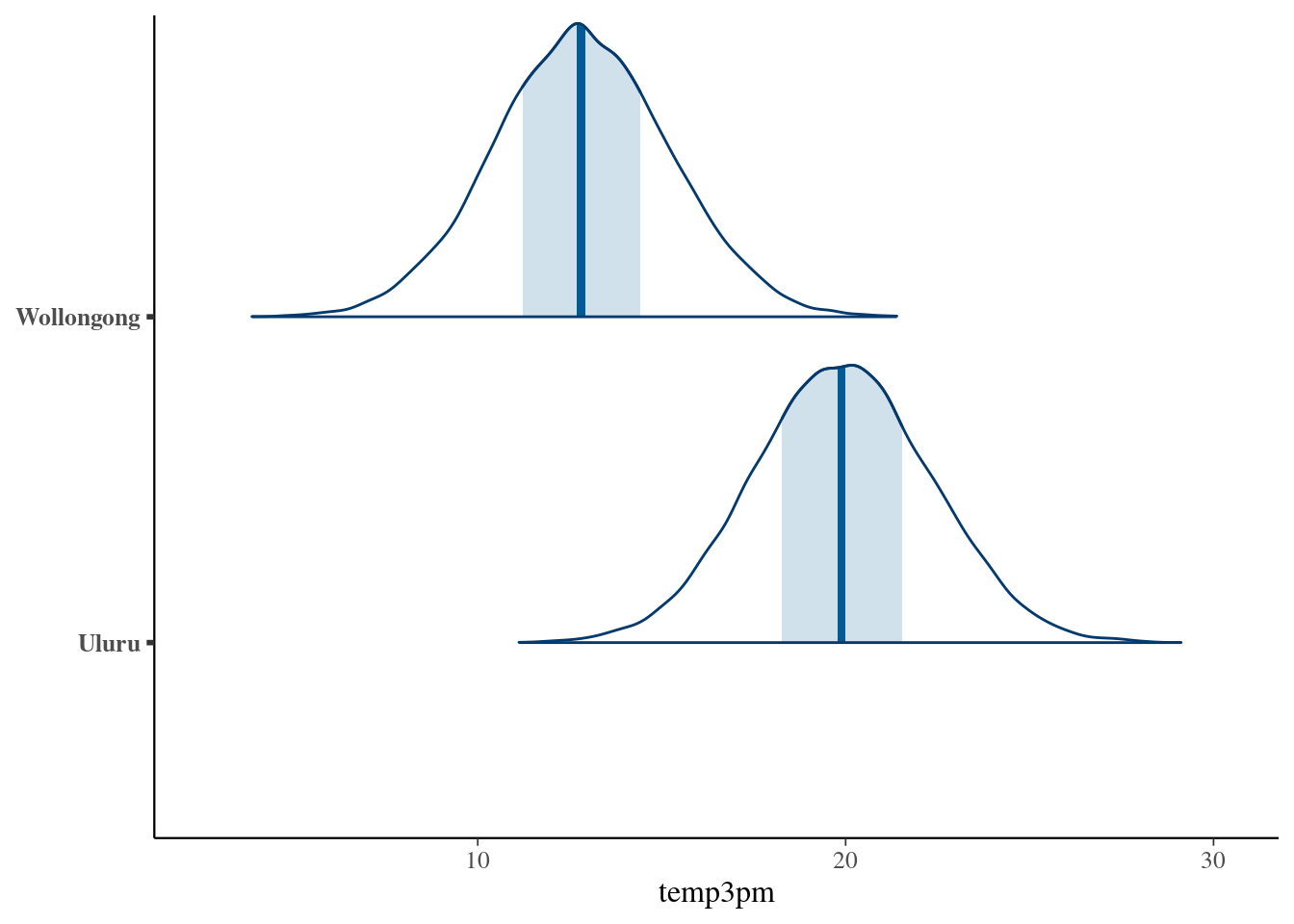 ## Optional: Utilizing interaction terms
## Optional: Utilizing interaction terms
Predictors interaction of 3 p.m. temperature (\(Y\)) with location (\(X_2\)) and 9 a.m. humidity (\(X_3\)).
ggplot(weather_WU, aes(y = temp3pm,
x = humidity9am,
color = location)) +
geom_point(size = 0.5) +
geom_smooth(method = "lm", se = FALSE)## `geom_smooth()` using formula = 'y ~ x'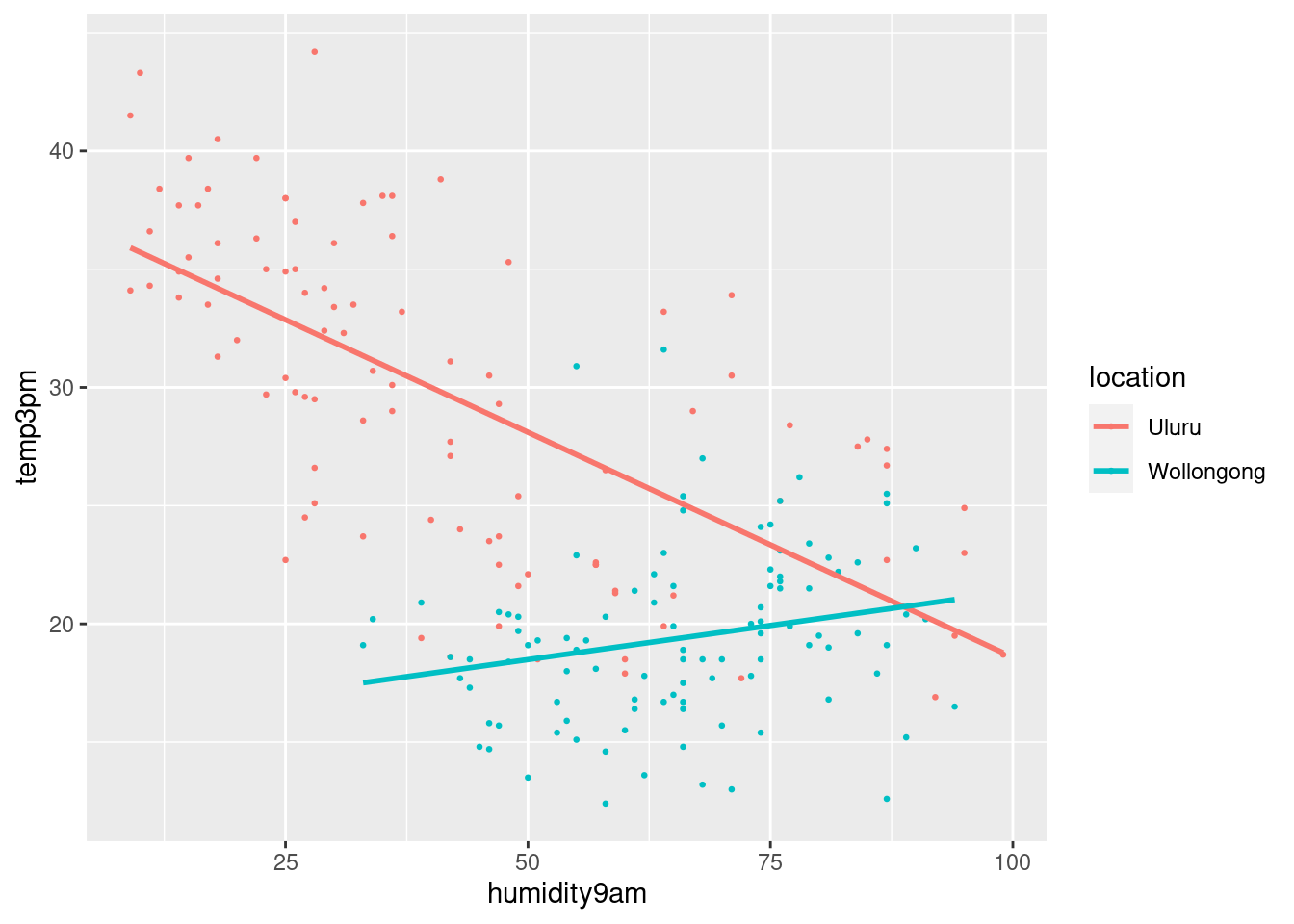
interaction_model <- stan_glm(
temp3pm ~ location + humidity9am + location:humidity9am,
data = weather_WU, family = gaussian,
prior_intercept = normal(25, 5),
prior = normal(0, 2.5, autoscale = TRUE),
prior_aux = exponential(1, autoscale = TRUE),
chains = 4, iter = 5000*2, seed = 84735)# Posterior summary statistics
tidy(interaction_model, effects = c("fixed", "aux"))## # A tibble: 6 × 3
## term estimate std.error
## <chr> <dbl> <dbl>
## 1 (Intercept) 37.6 0.909
## 2 locationWollongong -21.8 2.31
## 3 humidity9am -0.190 0.0193
## 4 locationWollongong:humidity9am 0.246 0.0372
## 5 sigma 4.47 0.229
## 6 mean_PPD 24.6 0.453Simulations:
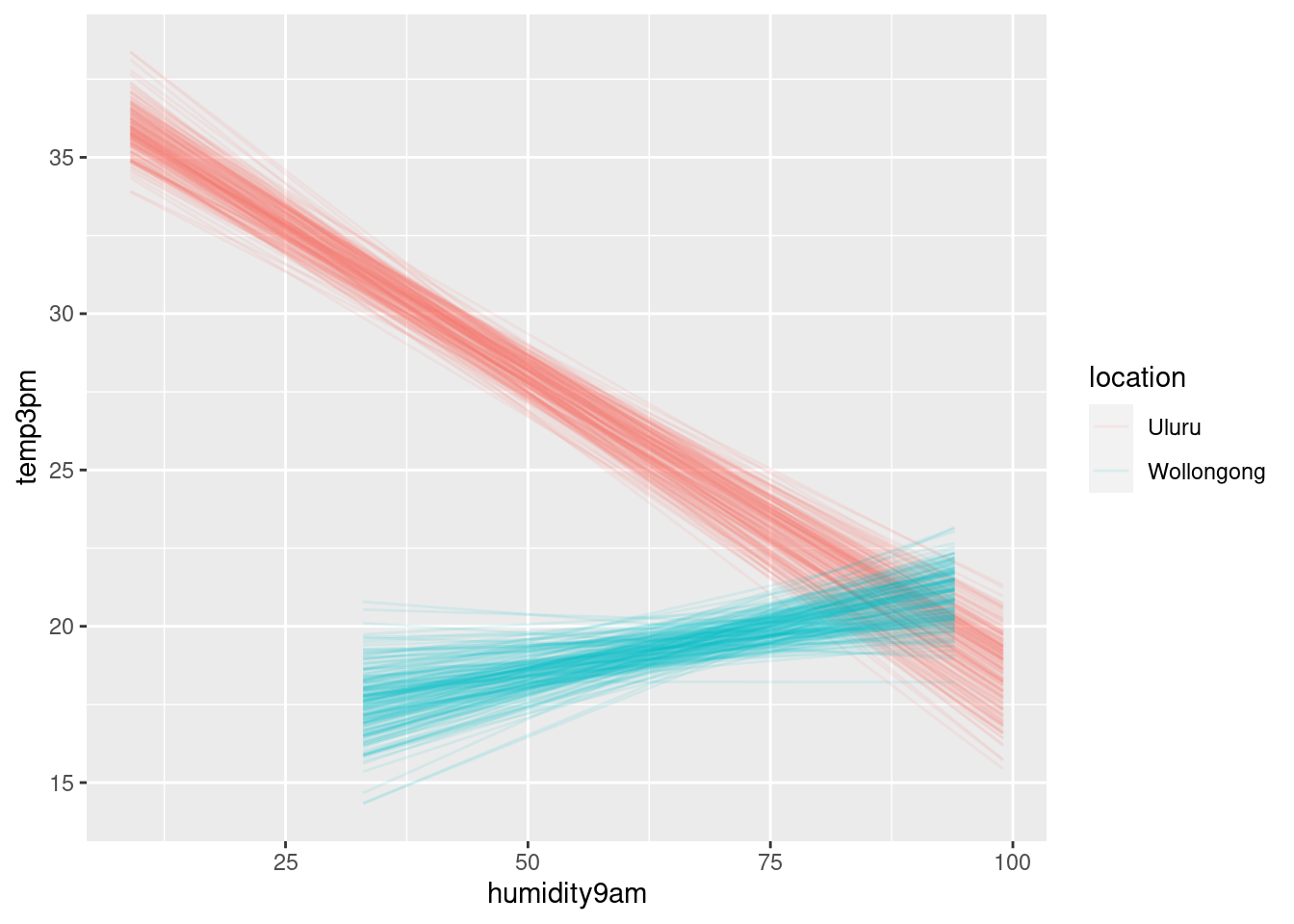
weather_WU %>%
add_fitted_draws(interaction_model, n = 200) %>%
ggplot(aes(x = humidity9am,
y = temp3pm,
color = location)) +
geom_line(aes(y = .value, group = paste(location, .draw)), alpha = 0.1)## Warning: `fitted_draws` and `add_fitted_draws` are deprecated as their names were confusing.
## - Use [add_]epred_draws() to get the expectation of the posterior predictive.
## - Use [add_]linpred_draws() to get the distribution of the linear predictor.
## - For example, you used [add_]fitted_draws(..., scale = "response"), which
## means you most likely want [add_]epred_draws(...).
## NOTE: When updating to the new functions, note that the `model` parameter is now
## named `object` and the `n` parameter is now named `ndraws`.