9.9 Using default rstanarm priors
Authors recommend using the default prior from rstanarm:
bike_model_default <- rstanarm::stan_glm(
rides ~ temp_feel, data = bikes,
family = gaussian,
# here very specific prior on sd
prior_intercept = normal(5000, 2.5, autoscale = TRUE), # <- see autoscale arg.
prior = normal(0, 2.5, autoscale = TRUE),
prior_aux = exponential(1, autoscale = TRUE),
chains = 4, iter = 5000*2, seed = 84735)prior_summary(bike_model_default)
Priors for model 'bike_model_default'
------
Intercept (after predictors centered)
Specified prior:
~ normal(location = 5000, scale = 2.5)
Adjusted prior:
~ normal(location = 5000, scale = 3937)
Coefficients
Specified prior:
~ normal(location = 0, scale = 2.5)
Adjusted prior:
~ normal(location = 0, scale = 351)
Auxiliary (sigma)
Specified prior:
~ exponential(rate = 1)
Adjusted prior:
~ exponential(rate = 0.00064)
------
See help('prior_summary.stanreg') for more detailsIt uses weakly informative priors using the scale of the data.
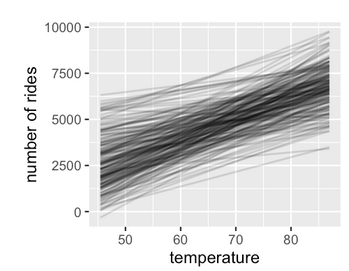
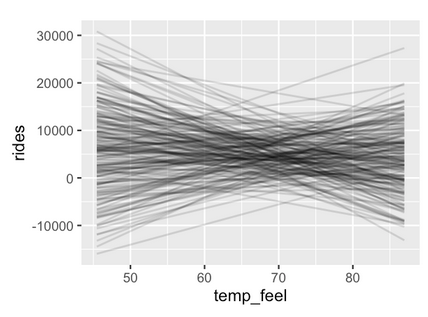
Figure 9.1: Fig 9.5 and 9.14 from Bayes Rules!