17.2 Hierarchical Model with varying intercept
17.2.1 Model buildings
17.2.1.1 Layer 1 Within-group: within runner
\[Y_{ij} | \beta_0, \beta_1, \sigma_j \sim N(\mu_{ij}, \sigma_j^2)\]
We added a bunch of \(j\)! So now we have runner specific mean (\(\mu_{ij}\)) and the variance with a runner (\(\sigma_j\))
\[\mu_{ij} = \beta_{0j} + \beta_1X_{ij}\] Here we are using a specific intercept for each runner (\(\beta_{oj}\)) but we are still using a global age coefficient (\(\beta_1\)).
17.2.1.2 Layer 2: Between Runners
Quizz!

Which of our current parameters (\(\beta_{0j}, \beta_1, \sigma_y\)) do we need to model in the next layer? (hint:title)
\[\beta_{0j} | \beta_{0}, \sigma_0 \overset{\text{ind}}{\sim} N(\beta_0, \sigma_0^2)\]
\(\beta_{0j}\) is our intercept for each runner and it follow a normal distribution with the global average of intercept (\(\beta_0\)) and the between-group variability (\(\sigma_0\)).
Now quiz!

For Which model parameters must we specify priors in the final layer of our hierarchical regression model?
\[\beta_{0c} \sim N(m_0, s_0^2)\]
\[\beta_1 \sim N(m_1, s_1^2)\]
\[\sigma_y \sim Exp(l_y)\]
\[\sigma_0 \sim Exp(l_0)\]
Normal hierarchical regression assumptions:
structure of the data: conditioned on \(X_{ij}\), \(Y_{ij}\) on any group j is independant of other group k but different data point within the same group are correlated
structure of the relationship: Linear relation
structure of variability within groups: Within any group j at any predictor value \(X_{ij}\) the observed values of \(Y_{ij}\) will vary normally
Structure of variability between groups
17.2.1.3 Tuning the prior
\[\beta_{0c} \sim N(100, 10^2)\] runing tine is around 80 - 120 mins
\[\beta_1 \sim N(2.5, 1^2)\] We just know that it increase and it can range from 0.5 to 4.5 mins / year (on average)
\[\sigma_y \sim Exp(0.078)\]
\[\sigma_0 \sim Exp(1)\]
Then we use weakly informative priors.
running_model_1_prior <- stan_glmer(
net ~ age + (1 | runner), # formula
data = running, family = gaussian,
prior_intercept = normal(100, 10),
prior = normal(2.5, 1),
prior_aux = exponential(1, autoscale = TRUE),
prior_covariance = decov(reg = 1, conc = 1, shape = 1, scale = 1),
chains = 4, iter = 5000*2, seed = 84735,
prior_PD = TRUE) # just the prior##
## SAMPLING FOR MODEL 'continuous' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 2.2e-05 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.22 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 10000 [ 0%] (Warmup)
## Chain 1: Iteration: 1000 / 10000 [ 10%] (Warmup)
## Chain 1: Iteration: 2000 / 10000 [ 20%] (Warmup)
## Chain 1: Iteration: 3000 / 10000 [ 30%] (Warmup)
## Chain 1: Iteration: 4000 / 10000 [ 40%] (Warmup)
## Chain 1: Iteration: 5000 / 10000 [ 50%] (Warmup)
## Chain 1: Iteration: 5001 / 10000 [ 50%] (Sampling)
## Chain 1: Iteration: 6000 / 10000 [ 60%] (Sampling)
## Chain 1: Iteration: 7000 / 10000 [ 70%] (Sampling)
## Chain 1: Iteration: 8000 / 10000 [ 80%] (Sampling)
## Chain 1: Iteration: 9000 / 10000 [ 90%] (Sampling)
## Chain 1: Iteration: 10000 / 10000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.568 seconds (Warm-up)
## Chain 1: 0.588 seconds (Sampling)
## Chain 1: 1.156 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL 'continuous' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 1.6e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.16 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 10000 [ 0%] (Warmup)
## Chain 2: Iteration: 1000 / 10000 [ 10%] (Warmup)
## Chain 2: Iteration: 2000 / 10000 [ 20%] (Warmup)
## Chain 2: Iteration: 3000 / 10000 [ 30%] (Warmup)
## Chain 2: Iteration: 4000 / 10000 [ 40%] (Warmup)
## Chain 2: Iteration: 5000 / 10000 [ 50%] (Warmup)
## Chain 2: Iteration: 5001 / 10000 [ 50%] (Sampling)
## Chain 2: Iteration: 6000 / 10000 [ 60%] (Sampling)
## Chain 2: Iteration: 7000 / 10000 [ 70%] (Sampling)
## Chain 2: Iteration: 8000 / 10000 [ 80%] (Sampling)
## Chain 2: Iteration: 9000 / 10000 [ 90%] (Sampling)
## Chain 2: Iteration: 10000 / 10000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.562 seconds (Warm-up)
## Chain 2: 0.593 seconds (Sampling)
## Chain 2: 1.155 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL 'continuous' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 1.5e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.15 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 10000 [ 0%] (Warmup)
## Chain 3: Iteration: 1000 / 10000 [ 10%] (Warmup)
## Chain 3: Iteration: 2000 / 10000 [ 20%] (Warmup)
## Chain 3: Iteration: 3000 / 10000 [ 30%] (Warmup)
## Chain 3: Iteration: 4000 / 10000 [ 40%] (Warmup)
## Chain 3: Iteration: 5000 / 10000 [ 50%] (Warmup)
## Chain 3: Iteration: 5001 / 10000 [ 50%] (Sampling)
## Chain 3: Iteration: 6000 / 10000 [ 60%] (Sampling)
## Chain 3: Iteration: 7000 / 10000 [ 70%] (Sampling)
## Chain 3: Iteration: 8000 / 10000 [ 80%] (Sampling)
## Chain 3: Iteration: 9000 / 10000 [ 90%] (Sampling)
## Chain 3: Iteration: 10000 / 10000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.571 seconds (Warm-up)
## Chain 3: 0.594 seconds (Sampling)
## Chain 3: 1.165 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL 'continuous' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 1.5e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.15 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 10000 [ 0%] (Warmup)
## Chain 4: Iteration: 1000 / 10000 [ 10%] (Warmup)
## Chain 4: Iteration: 2000 / 10000 [ 20%] (Warmup)
## Chain 4: Iteration: 3000 / 10000 [ 30%] (Warmup)
## Chain 4: Iteration: 4000 / 10000 [ 40%] (Warmup)
## Chain 4: Iteration: 5000 / 10000 [ 50%] (Warmup)
## Chain 4: Iteration: 5001 / 10000 [ 50%] (Sampling)
## Chain 4: Iteration: 6000 / 10000 [ 60%] (Sampling)
## Chain 4: Iteration: 7000 / 10000 [ 70%] (Sampling)
## Chain 4: Iteration: 8000 / 10000 [ 80%] (Sampling)
## Chain 4: Iteration: 9000 / 10000 [ 90%] (Sampling)
## Chain 4: Iteration: 10000 / 10000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.564 seconds (Warm-up)
## Chain 4: 0.589 seconds (Sampling)
## Chain 4: 1.153 seconds (Total)
## Chain 4:running |>
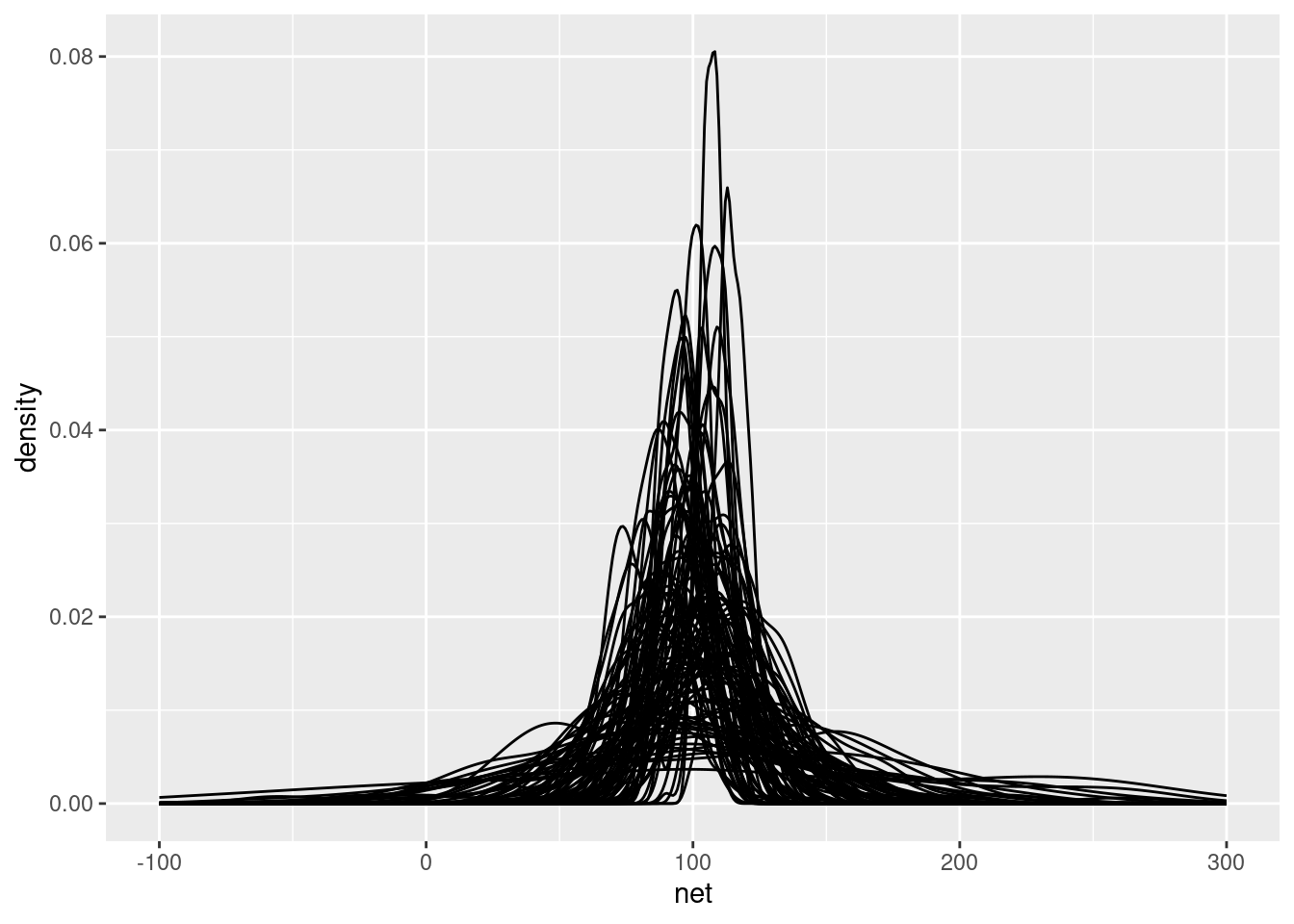
# here we just used 100 sims
add_predicted_draws(running_model_1_prior, n = 100) |>
ggplot(aes(x = net)) +
geom_density(aes(x = .prediction, group = .draw)) +
xlim(-100,300)## Warning:
## In add_predicted_draws(): The `n` argument is a deprecated alias for `ndraws`.
## Use the `ndraws` argument instead.
## See help("tidybayes-deprecated").## Warning: Removed 46 rows containing non-finite values (`stat_density()`).