9.5 Posterior simulatiion
Now we want to update our prior with data to get a posterior simulation!
# Load and plot data
library("bayesrules")
library("ggplot2")
data(bikes)
ggplot(bikes, aes(x = temp_feel, y = rides)) +
geom_point(size = 0.5) +
geom_smooth(formula = "y ~ x", method = "lm", se = FALSE) +
labs(title = "Washington DC Bike Ridership",
subtitle = "Fanaee-T, Hadi and Gama, Joao (2013)",
caption = "R4DS Book Club",
x = "what the temperature feels like (degrees Fahrenheit)",
y = "number of bikeshare rides") +
theme_minimal()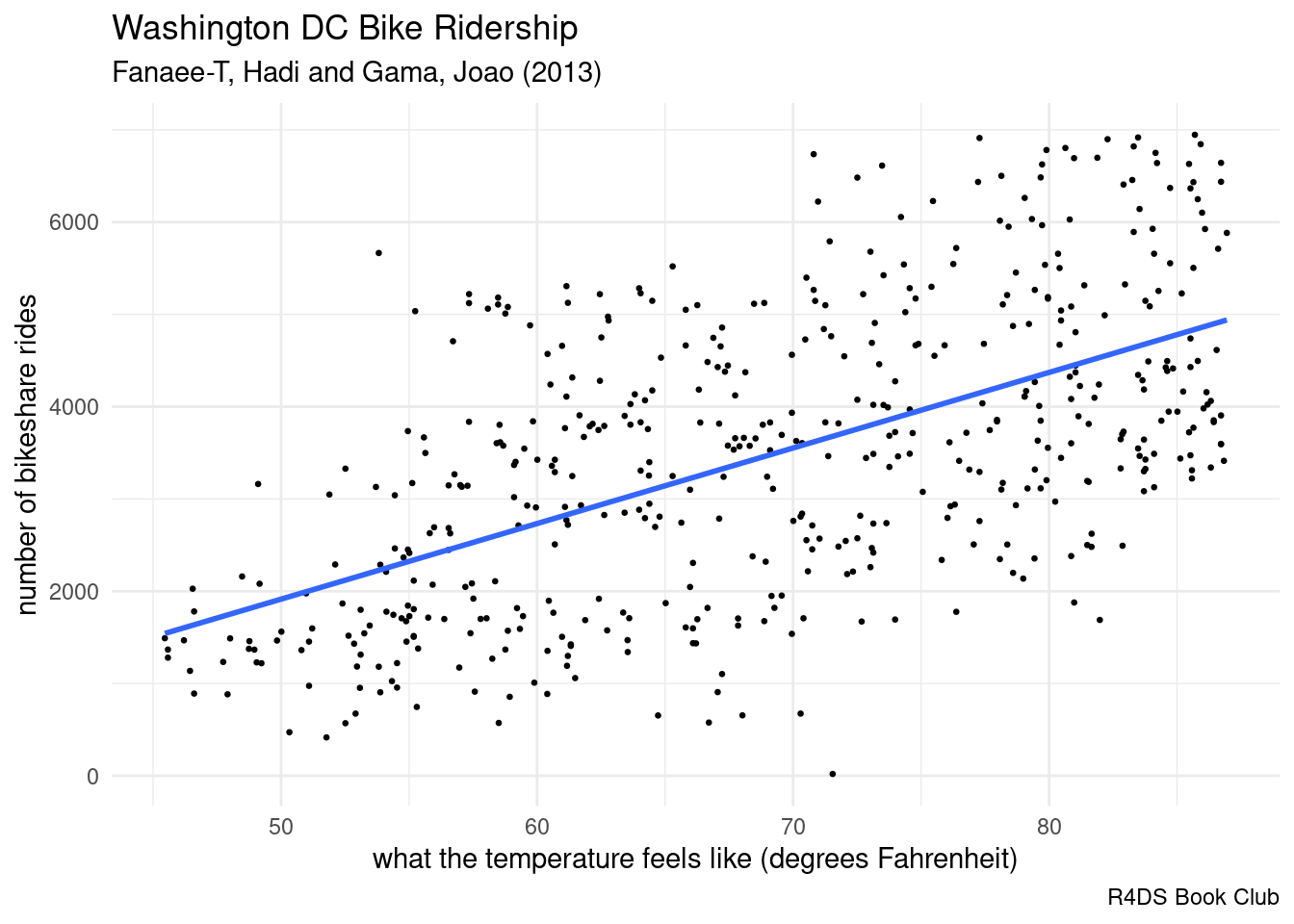
I am saving you from the triple integrals in the denominator of p 220 and we are jumping directly to MCMC!
We are going to use rstanarm : Rstan + arm = applied regression models
9.5.1 Simulation via rstanarm
bike_model <- rstanarm::stan_glm(
# data information
rides ~ temp_feel, # <- formula syntax (as used by lm, glm etc)
data = bikes,
family = gaussian, # <- we assume normal data
# Priors
prior_intercept = normal(5000, 1000), # centered intercept
prior = normal(100, 40),
prior_aux = exponential(0.0008), #notice the aux for auxiliary more after
# MCMC information
chains = 4, iter = 5000*2, seed = 84735)9.5.2 Simulation directly with rstan
# STEP 1: DEFINE the model
stan_bike_model <- "
data {
int<lower = 0> n;
vector[n] Y;
vector[n] X;
}
parameters {
real beta0;
real beta1;
real<lower = 0> sigma;
}
model {
Y ~ normal(beta0 + beta1 * X, sigma);
beta0 ~ normal(-2000, 1000);
beta1 ~ normal(100, 40);
sigma ~ exponential(0.0008);
}
"
# STEP 2: SIMULATE the posterior
stan_bike_sim <-
rstan::stan(model_code = stan_bike_model,
# data is structured a bit differently
data = list(n = nrow(bikes), Y = bikes$rides, X = bikes$temp_feel),
# same MCMC
chains = 4, iter = 5000*2, seed = 84735)The model will return 5000 x nb of chains simulation of our parameters. Rstanarm will change their name:
\(\beta_{0} \rightarrow (intercept)\)
- Note that this is \(\beta_{0}\) even though prior is specified for \(\beta_{0c}\)
\(\beta_{1} \rightarrow temp\_feel\)
But \(\sigma\) stays as \(\sigma\)
We need to check if the simulation went well for that we can?
Answer:
[X] Check the effective sample size: neff_ratio
[X] Compute rhat
[X] Examine trace plots
[X] Overlay the density plots of chains