6.6 Beta-Binomial MCMC
# define model in stan language
bb_model <- "
data {
int<lower = 0, upper = 10> Y;
}
parameters {
real<lower = 0, upper = 1> pi;
}
model {
Y ~ binomial(10, pi);
pi ~ beta(2, 2);
}
"
# https://github.com/stan-dev/rstan/wiki/Configuring-C---Toolchain-for-Windows#r-42
# use stan to simulate posterior
bb_sim <- rstan::stan(model_code = bb_model, data = list(Y = 9),
chains = 4, iter = 5000*2, seed = 84735)##
## SAMPLING FOR MODEL '7b64678a3565f32e51f77686e11b9c04' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 9e-06 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.09 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 10000 [ 0%] (Warmup)
## Chain 1: Iteration: 1000 / 10000 [ 10%] (Warmup)
## Chain 1: Iteration: 2000 / 10000 [ 20%] (Warmup)
## Chain 1: Iteration: 3000 / 10000 [ 30%] (Warmup)
## Chain 1: Iteration: 4000 / 10000 [ 40%] (Warmup)
## Chain 1: Iteration: 5000 / 10000 [ 50%] (Warmup)
## Chain 1: Iteration: 5001 / 10000 [ 50%] (Sampling)
## Chain 1: Iteration: 6000 / 10000 [ 60%] (Sampling)
## Chain 1: Iteration: 7000 / 10000 [ 70%] (Sampling)
## Chain 1: Iteration: 8000 / 10000 [ 80%] (Sampling)
## Chain 1: Iteration: 9000 / 10000 [ 90%] (Sampling)
## Chain 1: Iteration: 10000 / 10000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.040911 seconds (Warm-up)
## Chain 1: 0.042847 seconds (Sampling)
## Chain 1: 0.083758 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL '7b64678a3565f32e51f77686e11b9c04' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 5e-06 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.05 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 10000 [ 0%] (Warmup)
## Chain 2: Iteration: 1000 / 10000 [ 10%] (Warmup)
## Chain 2: Iteration: 2000 / 10000 [ 20%] (Warmup)
## Chain 2: Iteration: 3000 / 10000 [ 30%] (Warmup)
## Chain 2: Iteration: 4000 / 10000 [ 40%] (Warmup)
## Chain 2: Iteration: 5000 / 10000 [ 50%] (Warmup)
## Chain 2: Iteration: 5001 / 10000 [ 50%] (Sampling)
## Chain 2: Iteration: 6000 / 10000 [ 60%] (Sampling)
## Chain 2: Iteration: 7000 / 10000 [ 70%] (Sampling)
## Chain 2: Iteration: 8000 / 10000 [ 80%] (Sampling)
## Chain 2: Iteration: 9000 / 10000 [ 90%] (Sampling)
## Chain 2: Iteration: 10000 / 10000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.041655 seconds (Warm-up)
## Chain 2: 0.041915 seconds (Sampling)
## Chain 2: 0.08357 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL '7b64678a3565f32e51f77686e11b9c04' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 5e-06 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.05 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 10000 [ 0%] (Warmup)
## Chain 3: Iteration: 1000 / 10000 [ 10%] (Warmup)
## Chain 3: Iteration: 2000 / 10000 [ 20%] (Warmup)
## Chain 3: Iteration: 3000 / 10000 [ 30%] (Warmup)
## Chain 3: Iteration: 4000 / 10000 [ 40%] (Warmup)
## Chain 3: Iteration: 5000 / 10000 [ 50%] (Warmup)
## Chain 3: Iteration: 5001 / 10000 [ 50%] (Sampling)
## Chain 3: Iteration: 6000 / 10000 [ 60%] (Sampling)
## Chain 3: Iteration: 7000 / 10000 [ 70%] (Sampling)
## Chain 3: Iteration: 8000 / 10000 [ 80%] (Sampling)
## Chain 3: Iteration: 9000 / 10000 [ 90%] (Sampling)
## Chain 3: Iteration: 10000 / 10000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.041738 seconds (Warm-up)
## Chain 3: 0.040971 seconds (Sampling)
## Chain 3: 0.082709 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL '7b64678a3565f32e51f77686e11b9c04' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 5e-06 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.05 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 10000 [ 0%] (Warmup)
## Chain 4: Iteration: 1000 / 10000 [ 10%] (Warmup)
## Chain 4: Iteration: 2000 / 10000 [ 20%] (Warmup)
## Chain 4: Iteration: 3000 / 10000 [ 30%] (Warmup)
## Chain 4: Iteration: 4000 / 10000 [ 40%] (Warmup)
## Chain 4: Iteration: 5000 / 10000 [ 50%] (Warmup)
## Chain 4: Iteration: 5001 / 10000 [ 50%] (Sampling)
## Chain 4: Iteration: 6000 / 10000 [ 60%] (Sampling)
## Chain 4: Iteration: 7000 / 10000 [ 70%] (Sampling)
## Chain 4: Iteration: 8000 / 10000 [ 80%] (Sampling)
## Chain 4: Iteration: 9000 / 10000 [ 90%] (Sampling)
## Chain 4: Iteration: 10000 / 10000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.041935 seconds (Warm-up)
## Chain 4: 0.042458 seconds (Sampling)
## Chain 4: 0.084393 seconds (Total)
## Chain 4:Uses 4 chains and 10000 samples of which 1/2 are discarded by default for burn-in
Result is a stanfit object, which can be used to extract the samples
# for examining using view
chains <- as.data.frame(as.array(bb_sim, pars = "pi"))
# look at a zoom in of the sample trace
mcmc_trace(bb_sim, pars = "pi", window = c(50,100),size =0.1)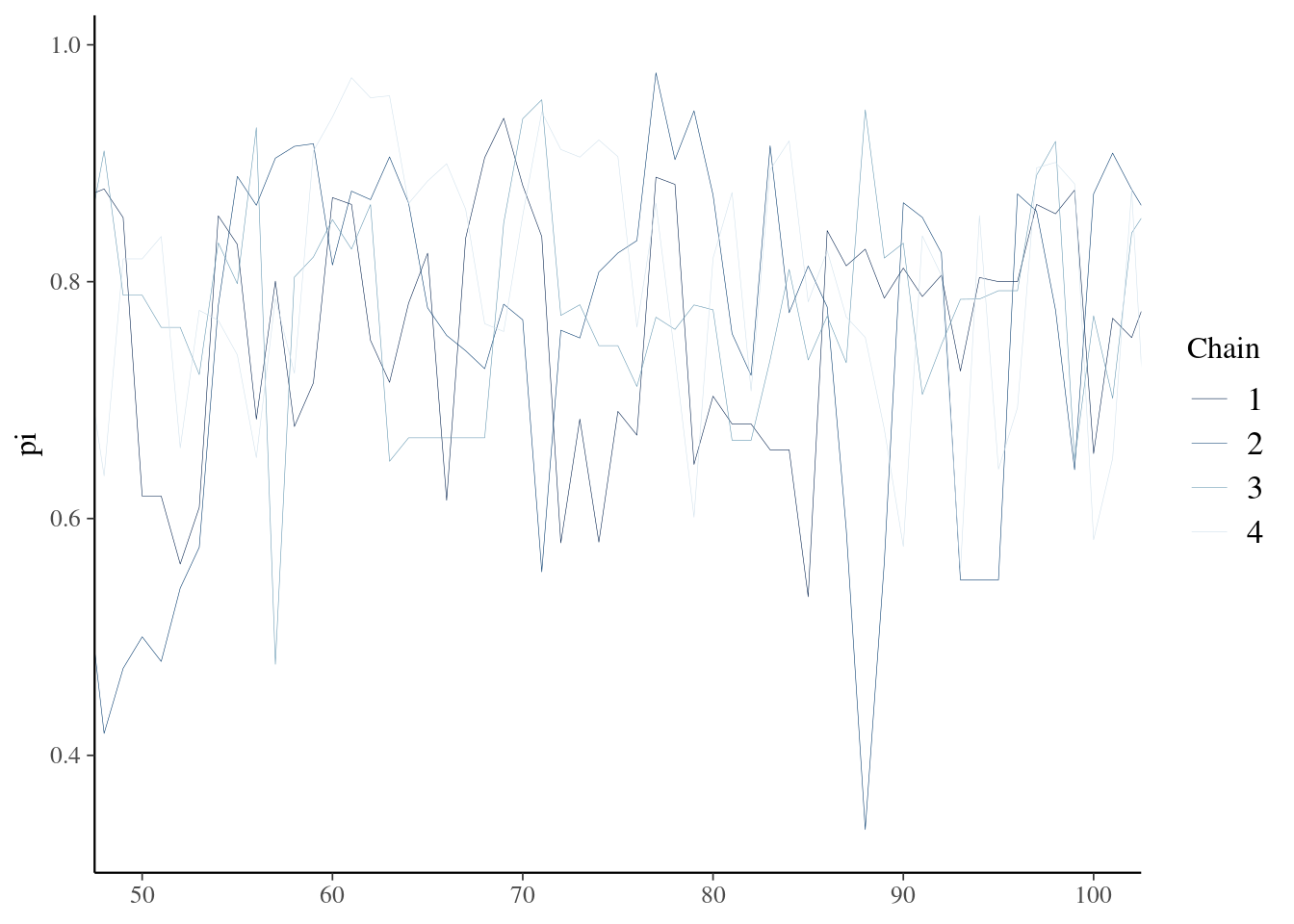
Trace shows the samples exploring the parameter space but also illustrates non-zero autocorrelation.
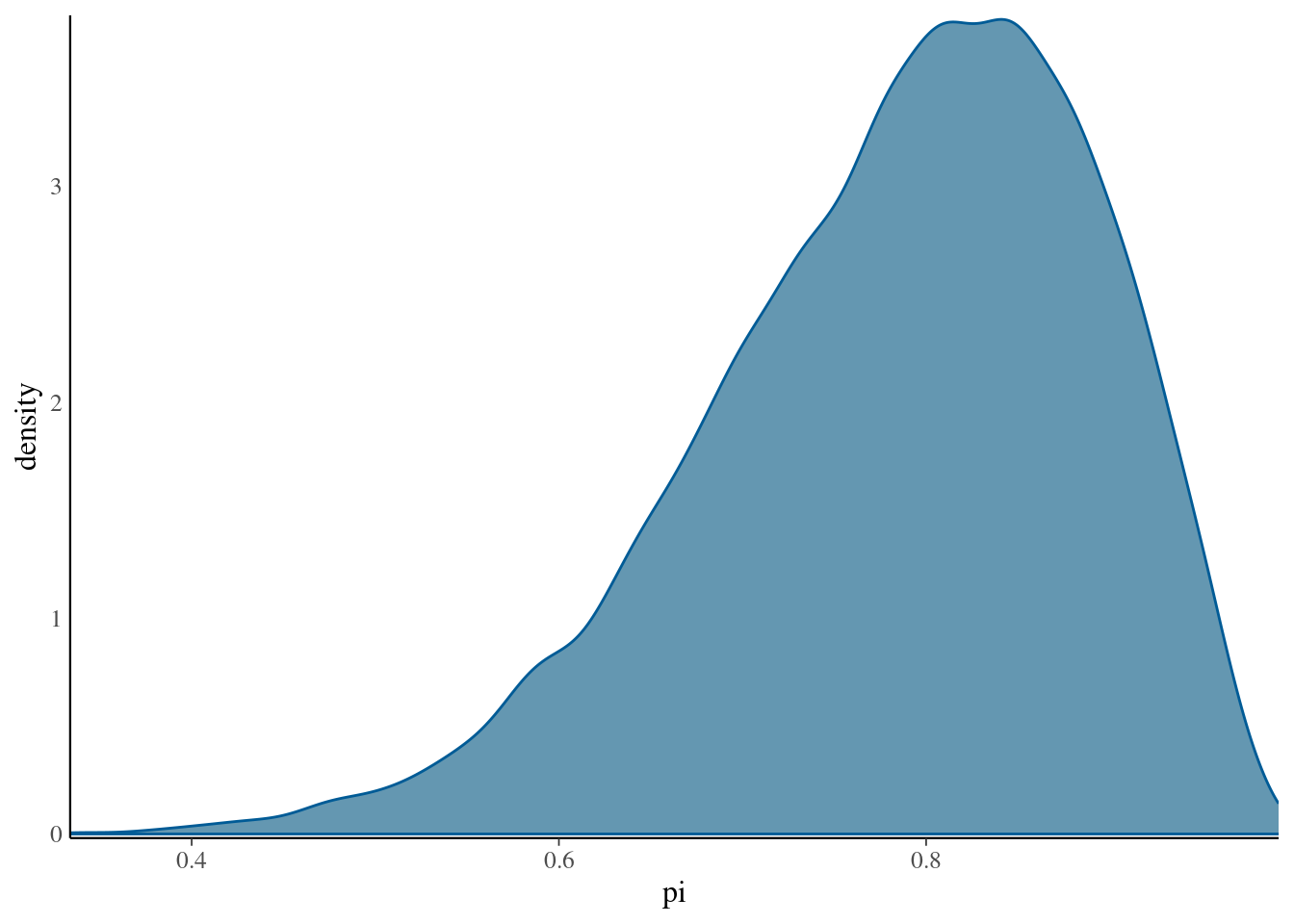
We can also plot the resulting distribution of samples (book shows that this is close to beta-binomial expected)
mcmc_dens(bb_sim, pars = "pi") +
yaxis_text(TRUE) +
ylab("density")