8.5 Posterior analysis with MCMC
Markov chains of \(\pi\) for 10,000 iterations each.
8.5.1 Posterior simulation
library(rstan)
# STEP 1: DEFINE the model
art_model <- "
data {
int<lower = 0, upper = 100> Y;
}
parameters {
real<lower = 0, upper = 1> pi;
}
model {
Y ~ binomial(100, pi);
pi ~ beta(4, 6);
}
"
# STEP 2: SIMULATE the posterior
art_sim <- stan(model_code = art_model,
data = list(Y = 14),
chains = 4,
iter = 5000*2,
seed = 84735)library(bayesplot)
# Parallel trace plots & density plots
mcmc_trace(art_sim, pars = "pi", size = 0.5) +
xlab("iteration")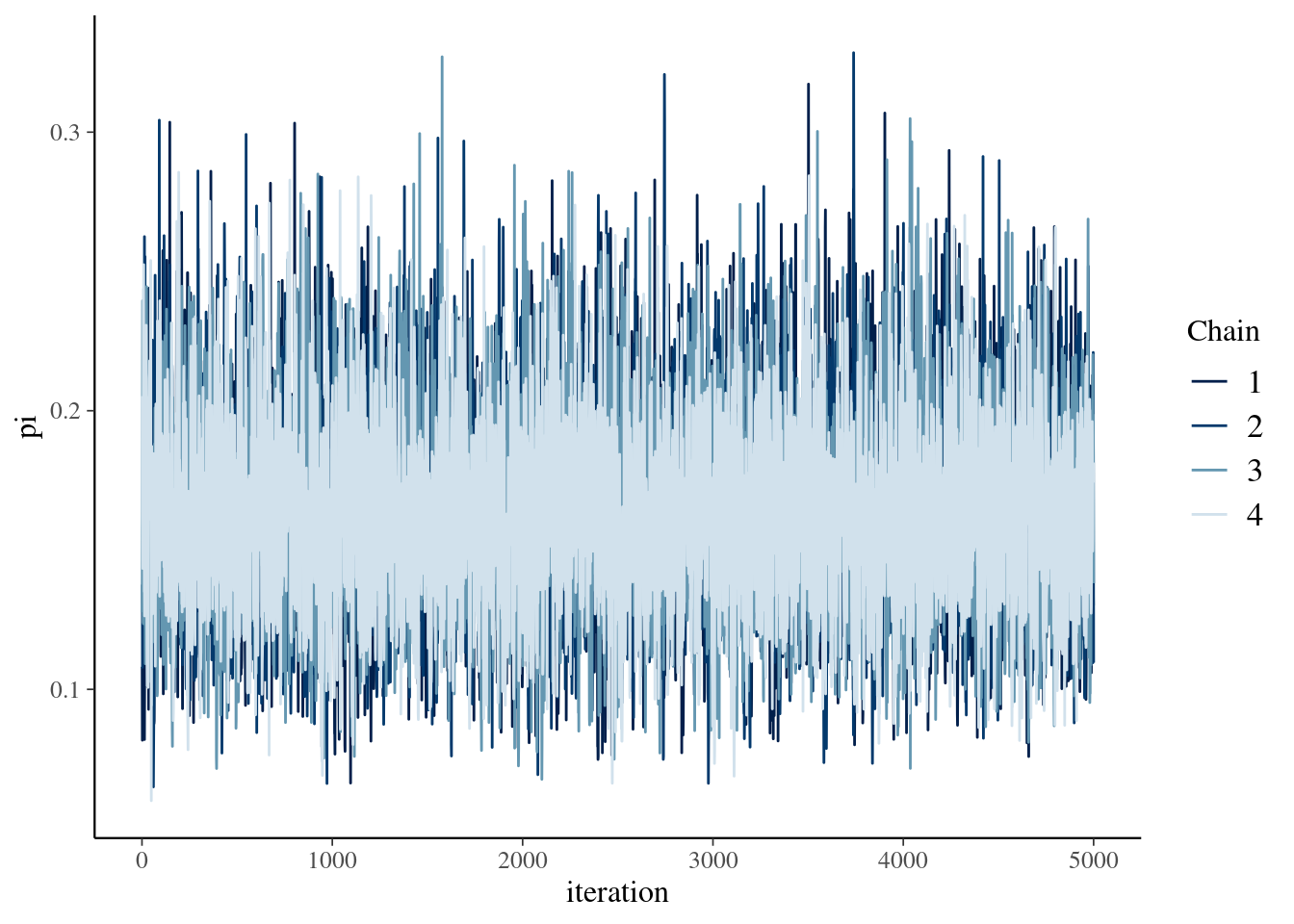
mcmc_dens_overlay(art_sim, pars = "pi")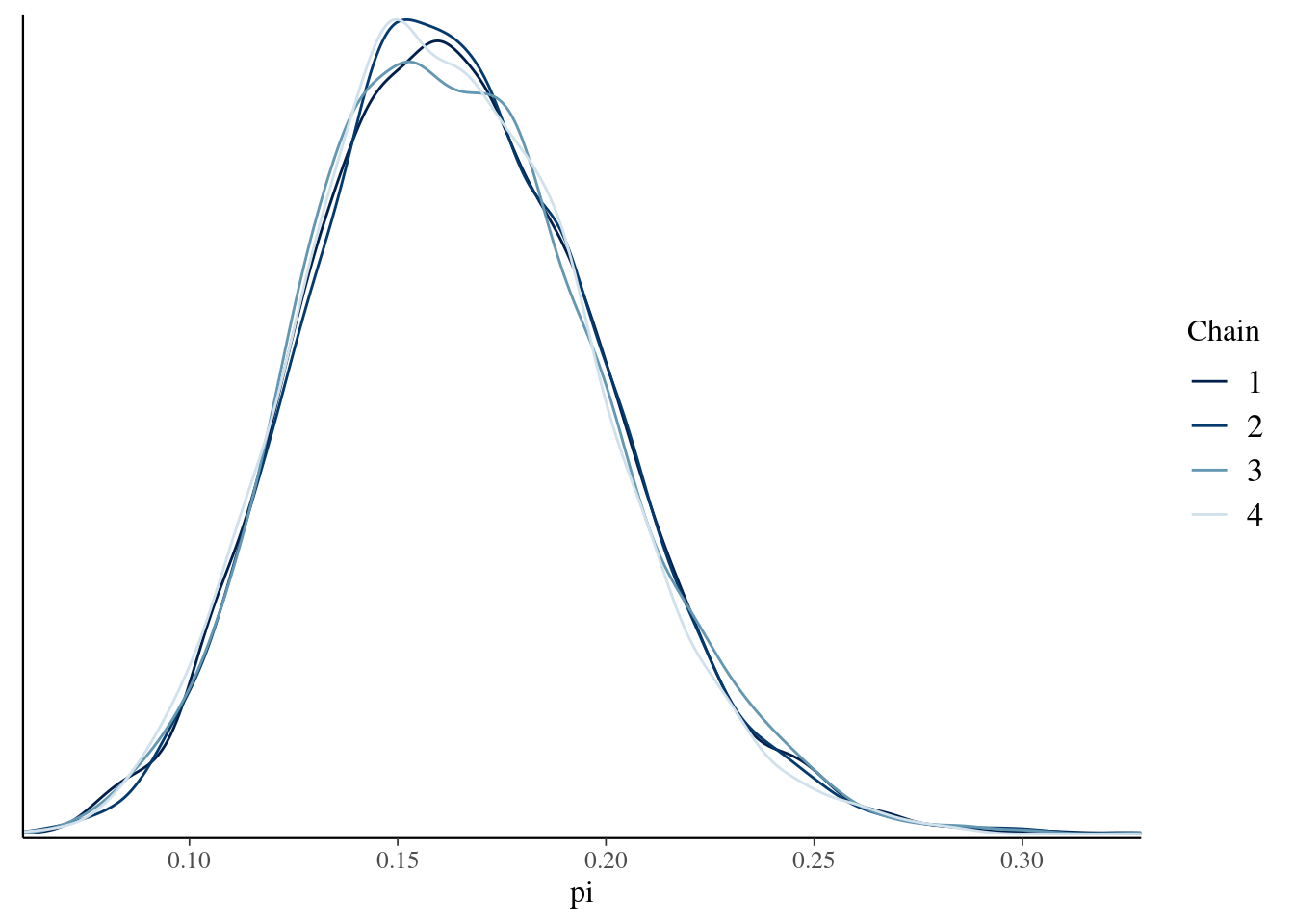
# Autocorrelation plot
mcmc_acf(art_sim, pars = "pi")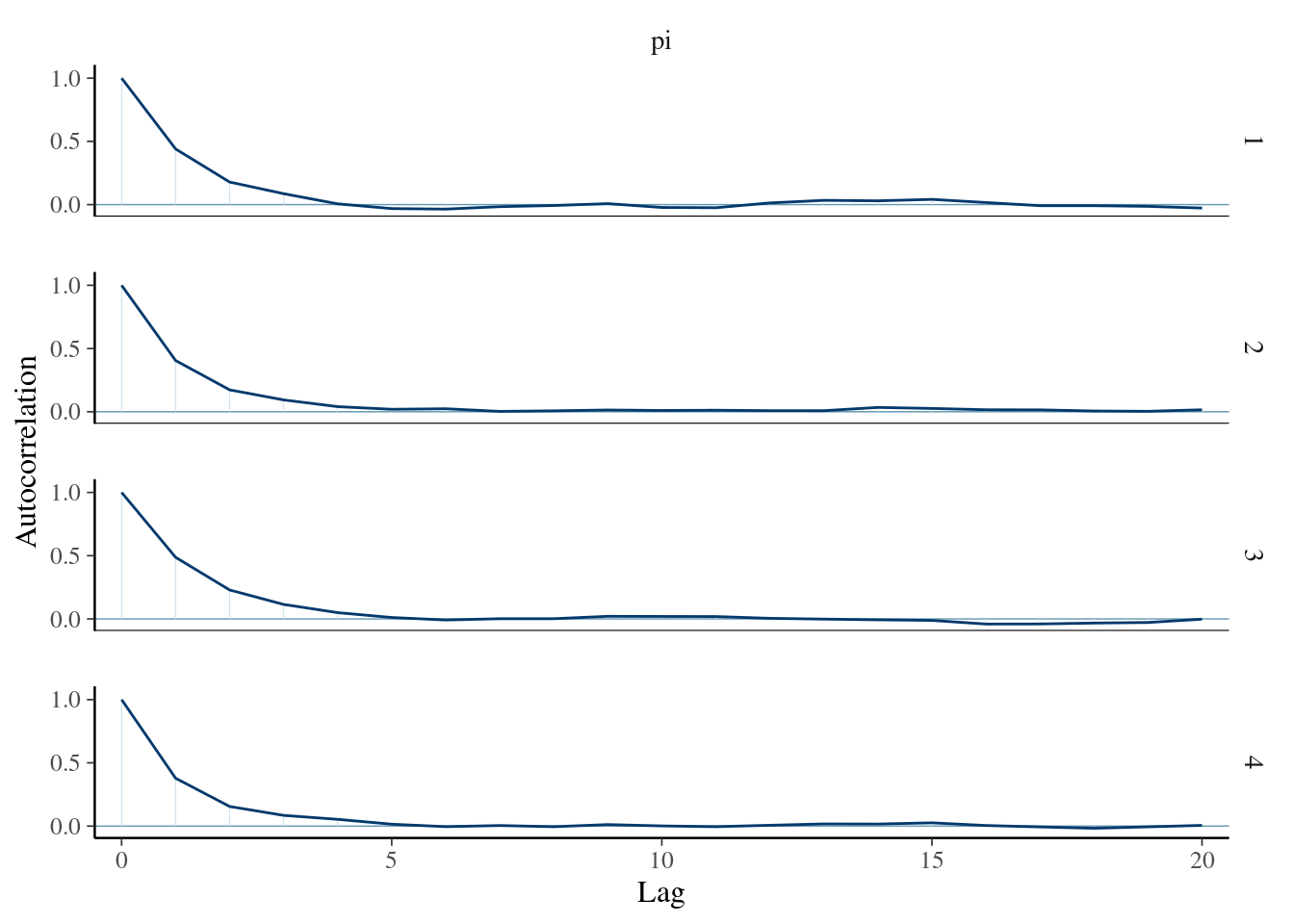
# Markov chain diagnostics
rhat(art_sim, pars = "pi")## [1] 1.000811neff_ratio(art_sim, pars = "pi")## [1] 0.3999558.5.2 Posterior estimation & hypothesis testing
Combined 20,000 Markov chain values.
# The actual Beta(18, 92) posterior
plot_beta(alpha = 18, beta = 92) +
lims(x = c(0, 0.35))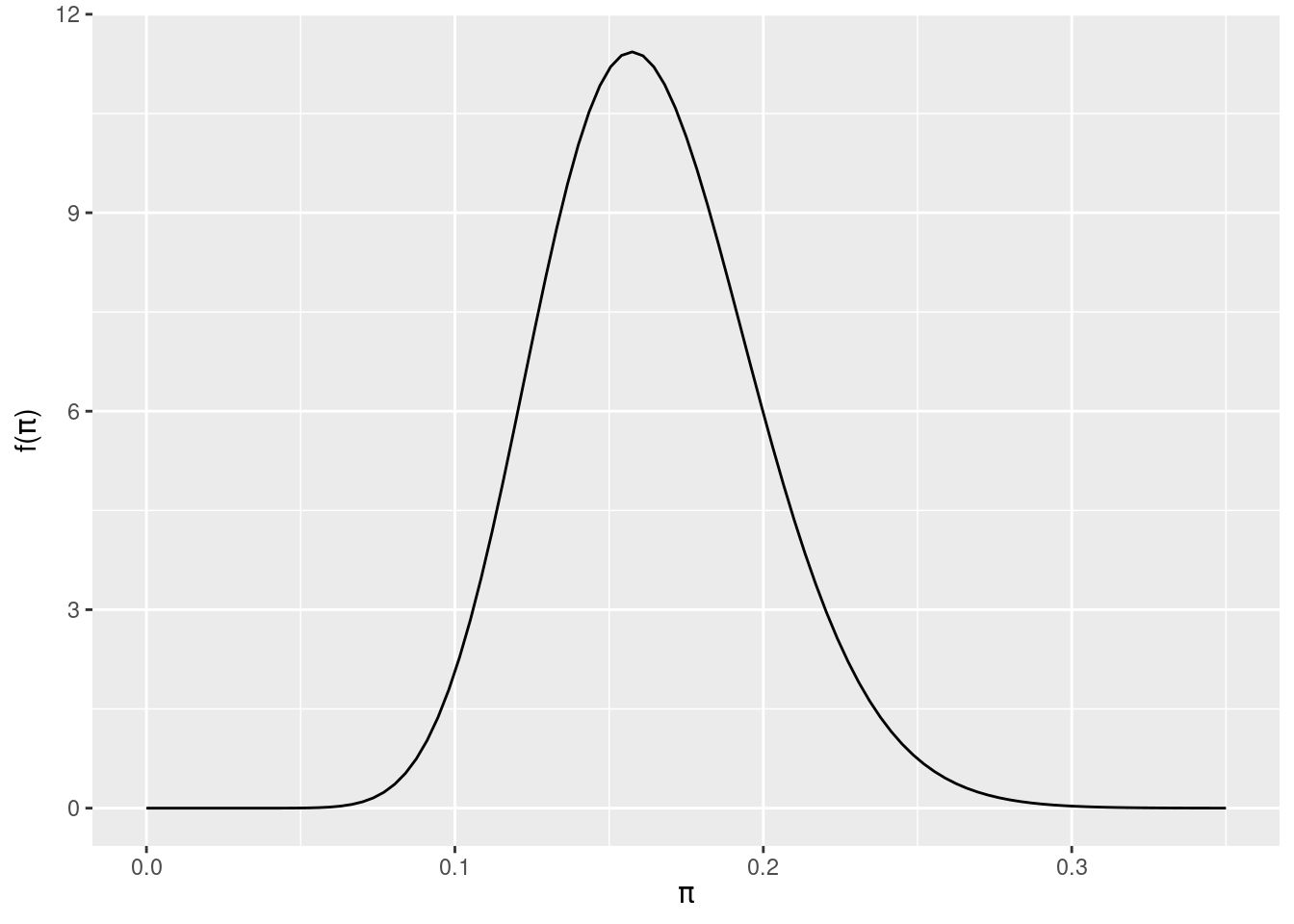
# MCMC posterior approximation
mcmc_dens(art_sim, pars = "pi") +
lims(x = c(0,0.35))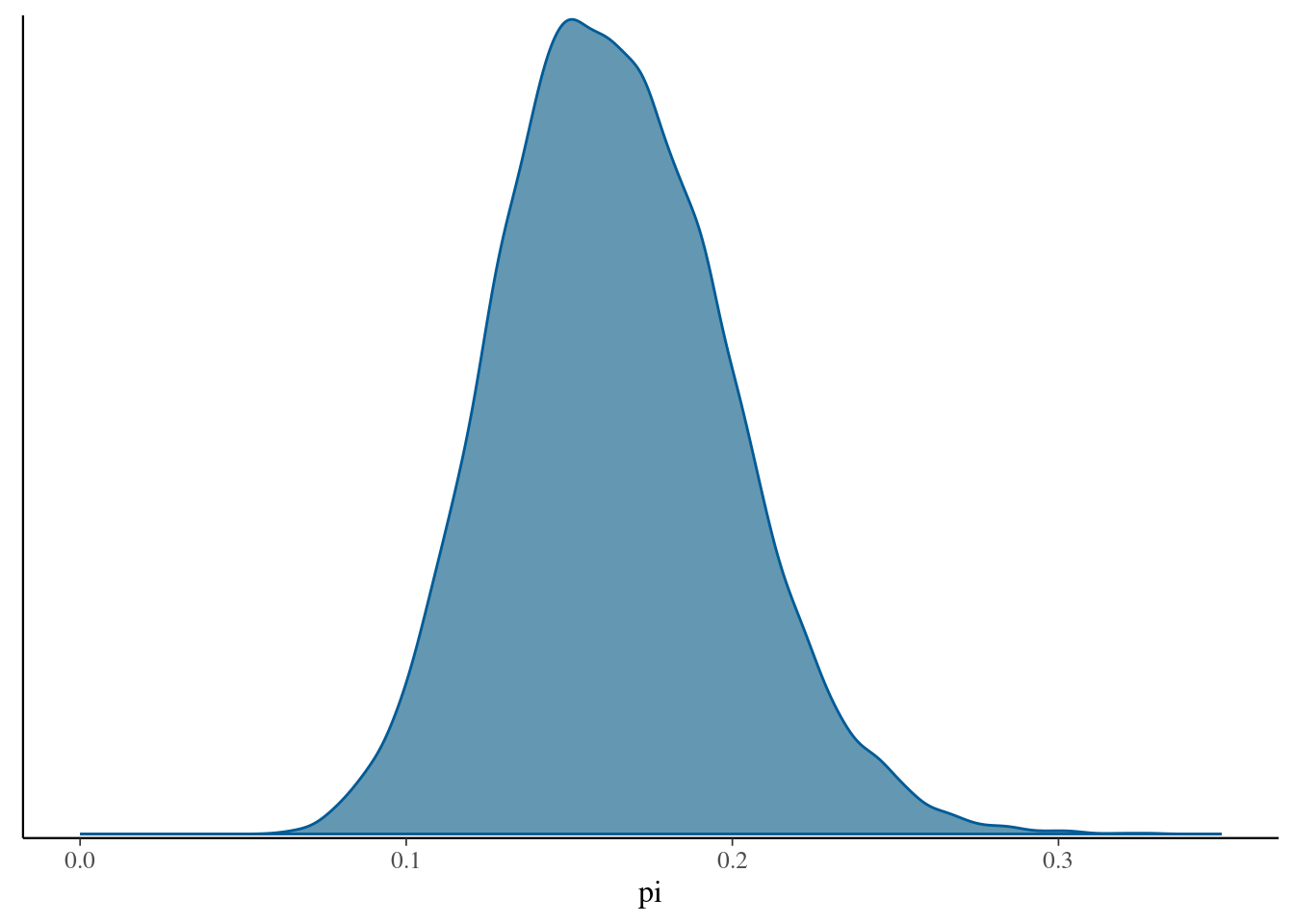
library(broom.mixed)
tidy(art_sim, conf.int = TRUE, conf.level = 0.95)## # A tibble: 1 × 5
## term estimate std.error conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 pi 0.162 0.0351 0.101 0.237# Shade in the middle 95% interval
mcmc_areas(art_sim, pars = "pi", prob = 0.95)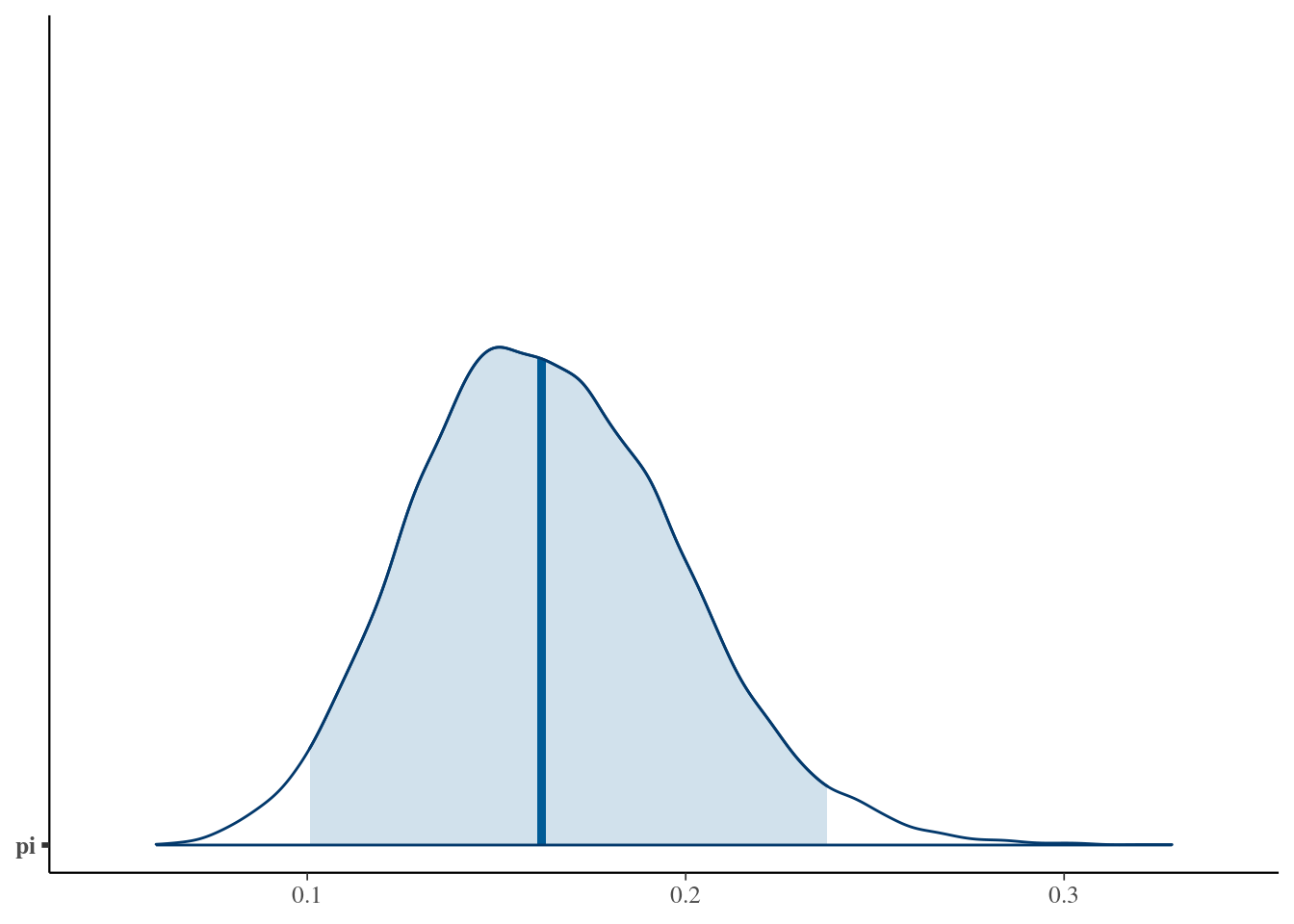
# Store the 4 chains in 1 data frame
art_chains_df <- as.data.frame(art_sim, pars = "lp__", include = FALSE)
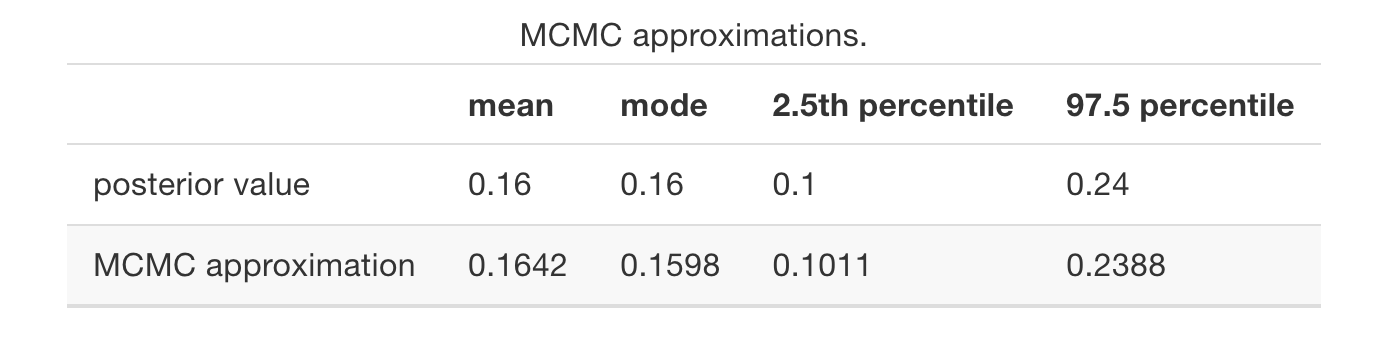
dim(art_chains_df)## [1] 20000 1# Calculate posterior summaries of pi
art_chains_df %>%
summarize(post_mean = mean(pi),
post_median = median(pi),
post_mode = sample_mode(pi),
lower_95 = quantile(pi, 0.025),
upper_95 = quantile(pi, 0.975))## post_mean post_median post_mode lower_95 upper_95
## 1 0.1637786 0.1618674 0.1507449 0.1006319 0.2374311Calculate summary statistics directly from the Markov chain values.
We can approximate the posterior probability \[P(\pi < 0.20 | Y =14)\]
library(janitor)
# Tabulate pi values that are below 0.20
art_chains_df %>%
mutate(exceeds = pi < 0.20) %>%
tabyl(exceeds)## exceeds n percent
## FALSE 3042 0.1521
## TRUE 16958 0.8479Compare results
8.5.3 Posterior prediction
Utilize the Markov chain values to approximate the posterior predictive model of \(Y'\). Repeating previous assumptions (sampling and posterior variability) and using rbinom(), we obtain:
# Set the seed
set.seed(1)
# Predict a value of Y' for each pi value in the chain
art_chains_df <- art_chains_df %>%
mutate(y_predict = rbinom(length(pi),
size = 20,
prob = pi))
# Check it out
art_chains_df %>%
head(3)## pi y_predict
## 1 0.10809936 1
## 2 0.08175123 1
## 3 0.09811126 2# Plot the 20,000 predictions
ggplot(art_chains_df, aes(x = y_predict)) +
stat_count()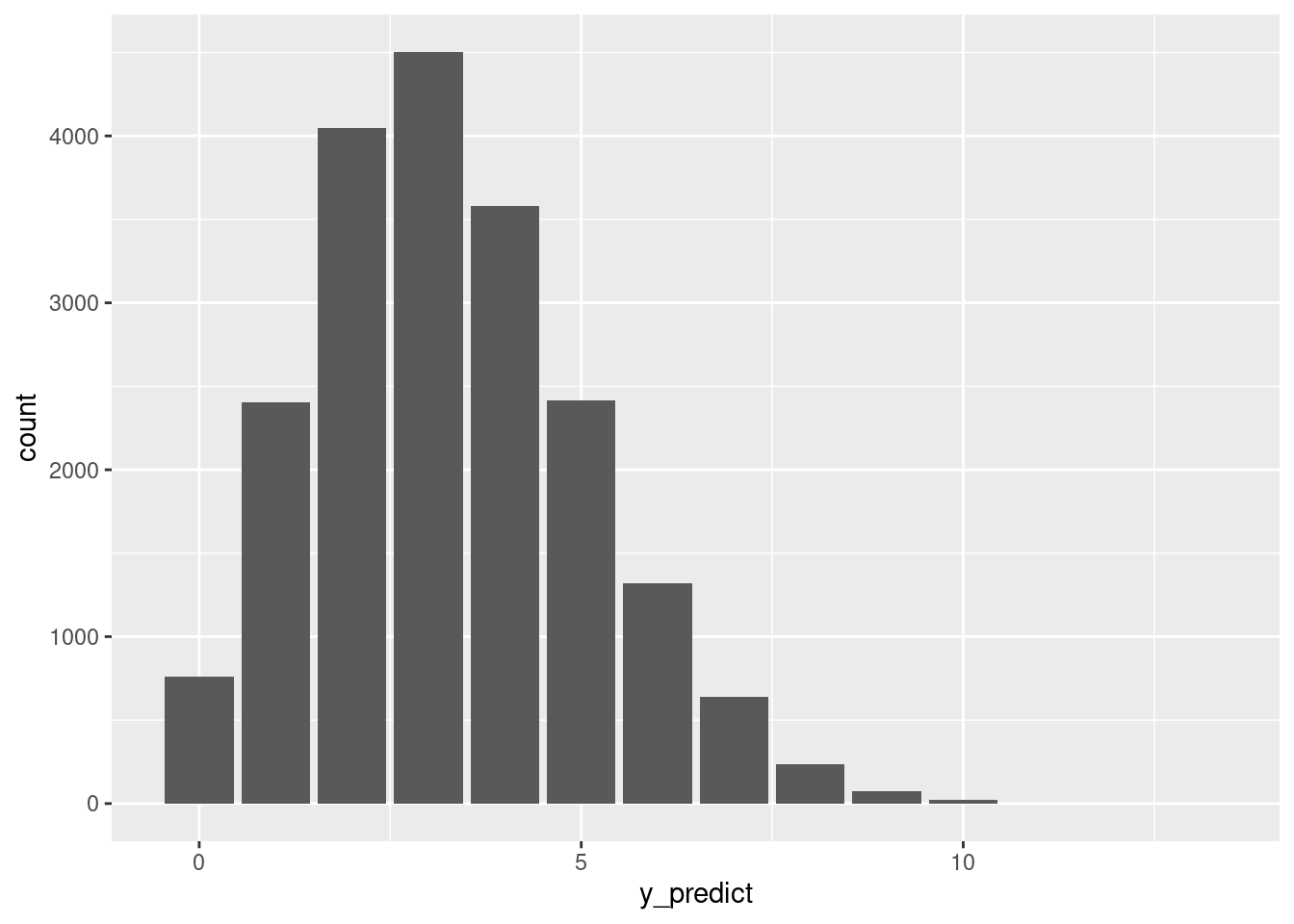
Approximate the posterior mean prediction \(E(Y'|Y=14)\) and the posterior prediction interval for \(Y'\).
art_chains_df %>%
summarize(mean = mean(y_predict),
lower_80 = quantile(y_predict, 0.1),
upper_80 = quantile(y_predict, 0.9))## mean lower_80 upper_80
## 1 3.2793 1 6