13.11 Replications
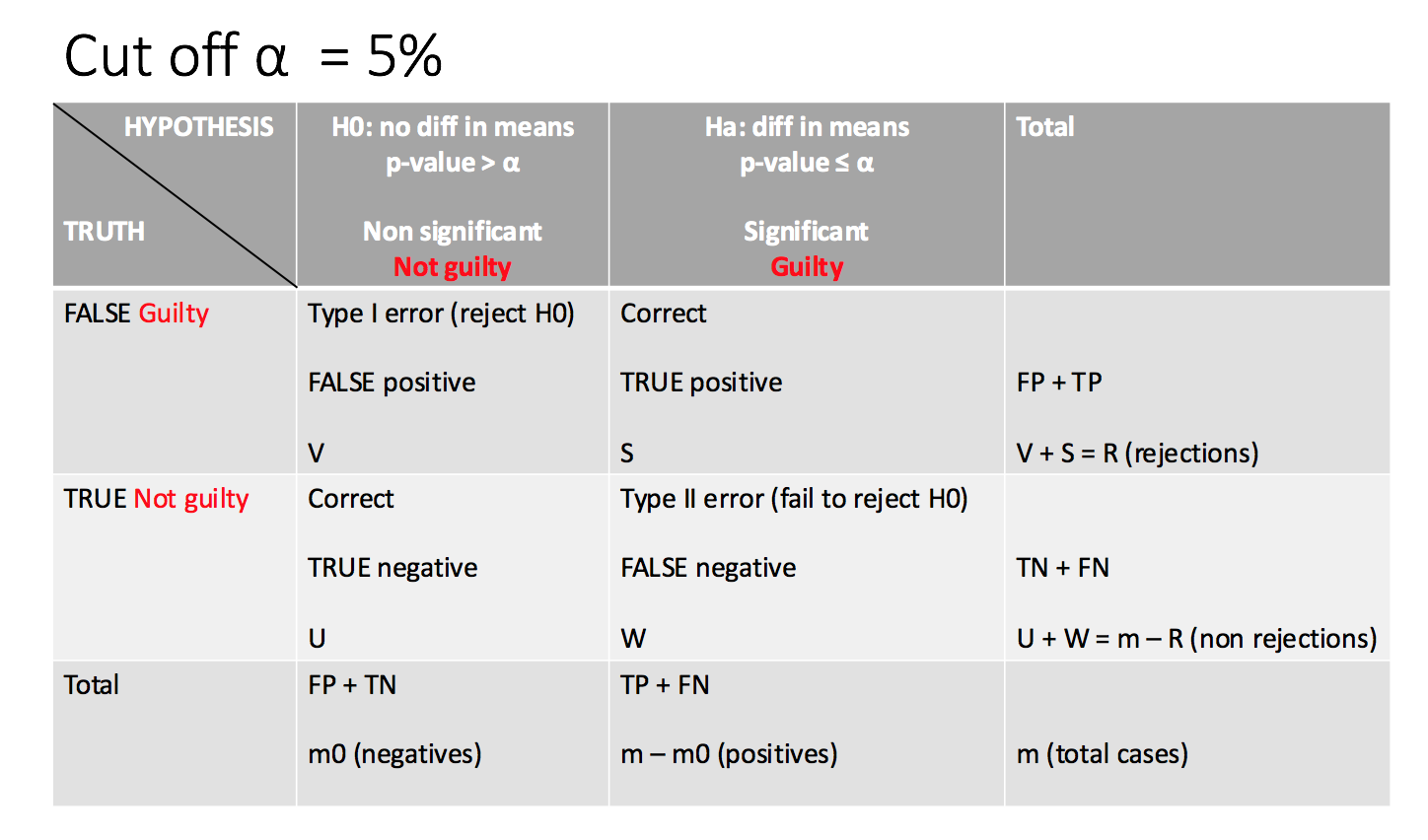
Little recap: There is a trade-off between type I & type II error
- Type I error reject \(H_0\) when \(H_0\) is TRUE
- Type I error Rate is the prob of type I error
- Type II error no reject \(H_0\) when \(H_0\) is FALSE
- POWER of hypothesis is the prob of not making type II error
credit: Federica Gazzelloni
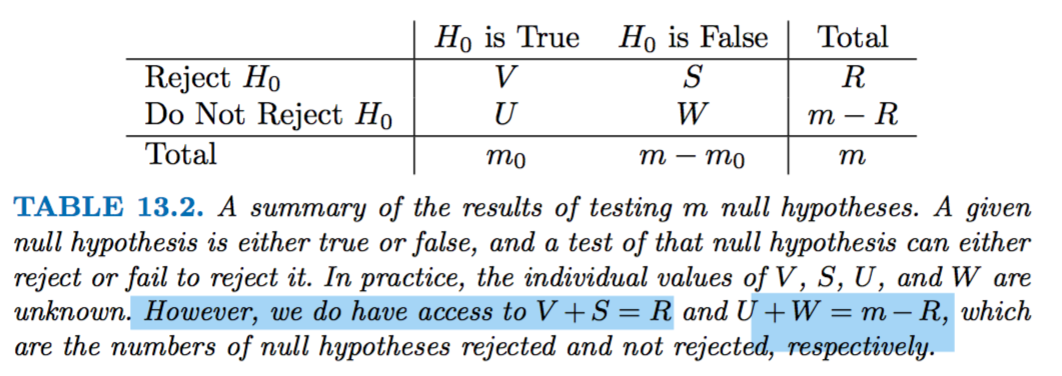
- R: sum of the number of pvalues which are below the threshold and will be rejected (n. rejections)
- m: total hypothesis testing
- m0: negatives
- m1: m-m0 positives
Now what we do is replicating the sample in a lab environment to obtain fake data, in order to do that we generate a matrix from a replication of normal distribution of the same size of our data.
n <- 24
m <- 8793
delta <- 2 # this is the sample split in two
positives <- 500 # m1
# negatives
m0<- m-positives # 8293
mat <- matrix(rnorm(n*m),m,n)
mat[1:positives,1:(n/2)] <- mat[1:positives,1:(n/2)] + deltaThen we do 1000 replications to see in what is the false discovery rate for this lab.
B<-1000
set.seed(1173)
results_global <- replicate(B,{
mat <- matrix(rnorm(n*m),m,n)
mat[1:positives,1:(n/2)] <- mat[1:positives,1:(n/2)] + delta
pvals = genefilter::rowttests(mat,null_hypothesis)$p.val
##Bonferroni
FP1 <- sum(pvals[-(1:positives)]<=0.05/m)
FN1 <- sum(pvals[1:positives]>0.05/m)
# p.adjust
qvals1 <- p.adjust(pvals,method = "fdr")
FP2<-sum(qvals1[-(1:positives)]<=0.05)
FN2 <- sum(qvals1[1:positives]>0.05)
# qvalue
qvals2 <- qvalue::qvalue(pvals)$qvalues
FP3<-sum(qvals2[-(1:positives)]<=0.05)
FN3 <- sum(qvals2[1:positives]>0.05)
c(FP1,FN1,FP2,FN2,FP3,FN3)
}) class(results_global)## [1] "matrix" "array"In this table are summarised the: false positives (FP) and the false negatives (FN) for three methods:
- Bonferroni
- FDR with p.adjust()
- FDR with qvalue()
The counts for FP and FN for the three cases:
## V1 V2 V3 V4 V5 V6
## FP1 0 0 0 0 0 0
## FN1 383 382 372 391 383 369
## FP2-padjust 22 22 20 32 22 20
## FN2-padjust 42 29 34 28 38 30
## FP3-qvalue 24 22 20 35 24 24
## FN3-qvalue 38 28 33 27 34 29The mean values or the proportions for the same methods:
## id Bonferroni FDR...padjust FDR...qvalue
## 1 FP 4.461594e-06 0.002773906 0.002960328
## 2 FN 7.645120e-01 0.081822000 0.077982000Benjamini-Hochberg
\[p(i)≤\frac{i}{m}\alpha\]
alpha <- 0.05
i = seq(along=pvals)
mypar(1,2)
plot(i,sort(pvals))
abline(0,i/m*alpha)
##close-up
plot(i[1:15],sort(pvals)[1:15],main="Close-up")
abline(0,i/m*alpha)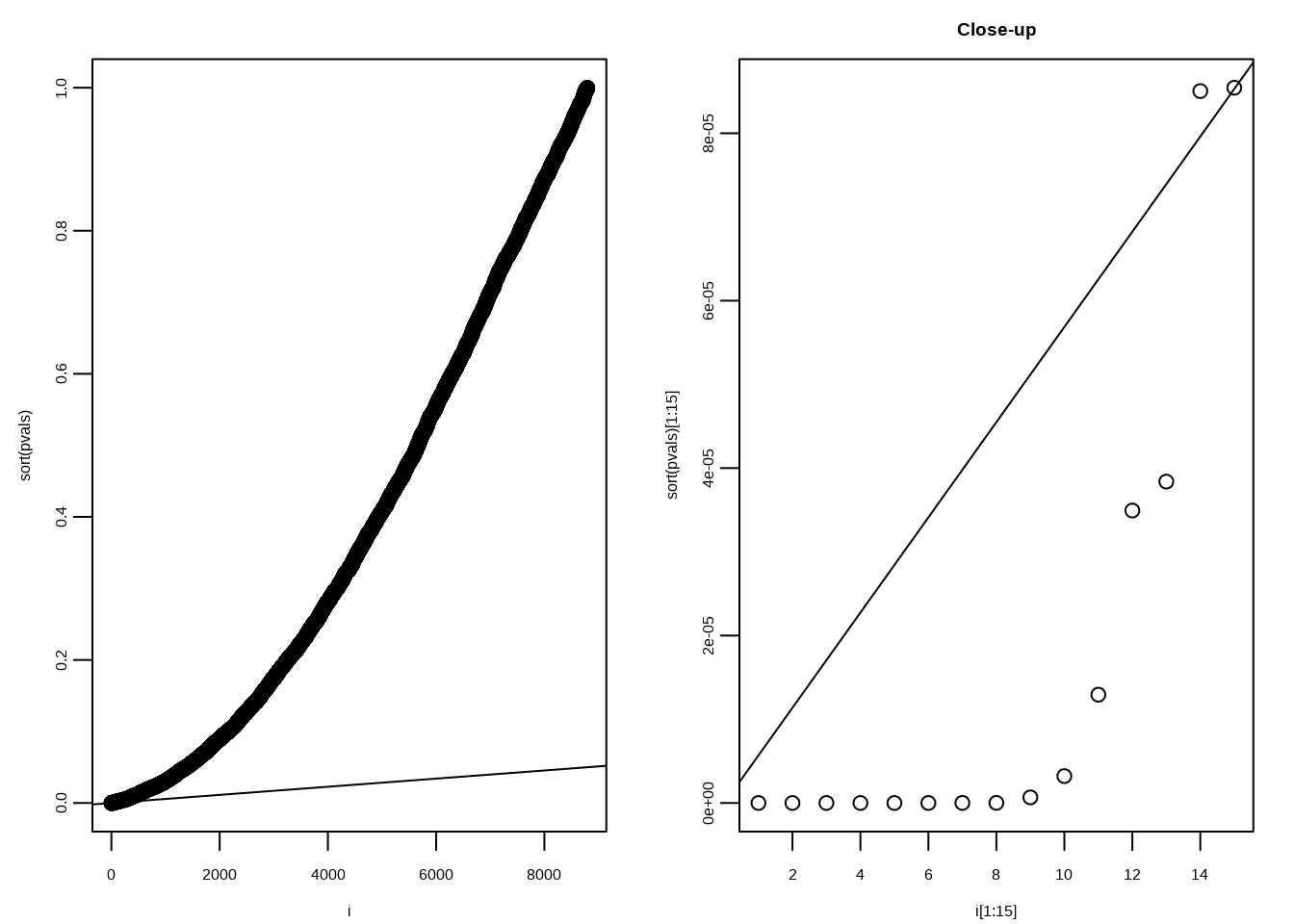
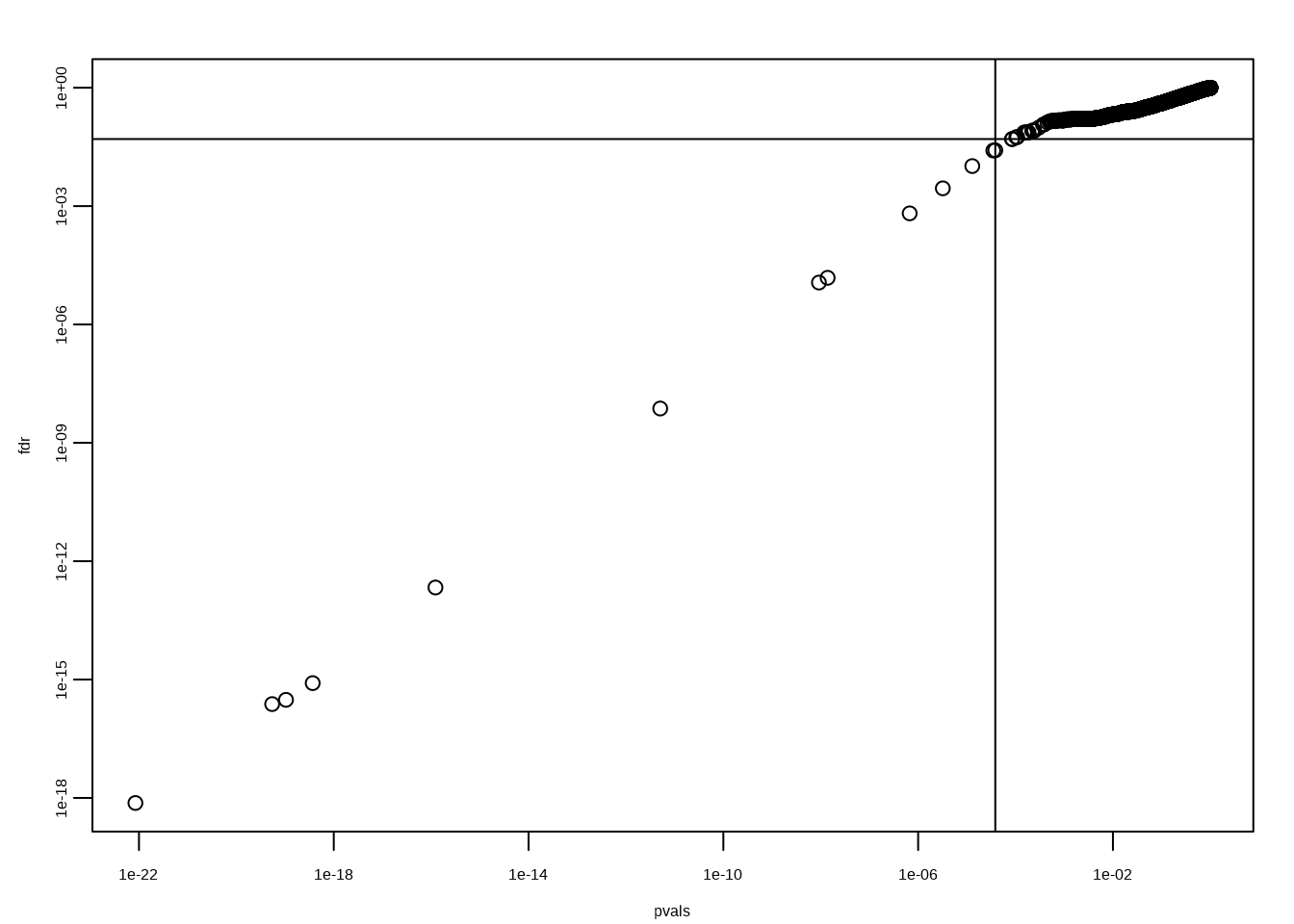
k <- max( which( sort(pvals) < i/m*alpha) )
cutoff <- sort(pvals)[k]
cat("k =",k,"p-value cutoff=",cutoff)## k = 13 p-value cutoff= 3.83879e-05fdr <- p.adjust(pvals, method="fdr")
mypar(1,1)
plot(pvals,fdr,log="xy")
abline(h=alpha,v=cutoff)