18.4 Factors and empty groups
18.4.1 Factors and empty groups
A final type of missingness is the empty group, a group that doesn’t contain any observations, which can arise when working with factors.
Here is a dataset that contains some health information about people.
health <- tibble::tibble(
name = c("Ikaia", "Oletta", "Leriah", "Dashay", "Tresaun"),
smoker = factor(c("no", "no", "no", "no", "no"),
levels = c("yes", "no")),
age = c(34, 88, 75, 47, 56),
)We want to count the number of smokers and non-smokers with dplyr::count() but it only gives us the amount of smokers because the group of smokers is empty
## # A tibble: 1 × 2
## smoker n
## <fct> <int>
## 1 no 5We can request count() to keep all the groups, even those not seen in the data by using .drop = FALSE:
## # A tibble: 2 × 2
## smoker n
## <fct> <int>
## 1 yes 0
## 2 no 5The same principle applies to ggplot2’s discrete axes, which will also drop levels that don’t have any values. You can force them to display by supplying drop = FALSE to the appropriate discrete axis
ggplot2::ggplot(
data = health,
mapping = ggplot2::aes(
x = .data[["smoker"]])
) +
ggplot2::geom_bar() +
ggplot2::scale_x_discrete()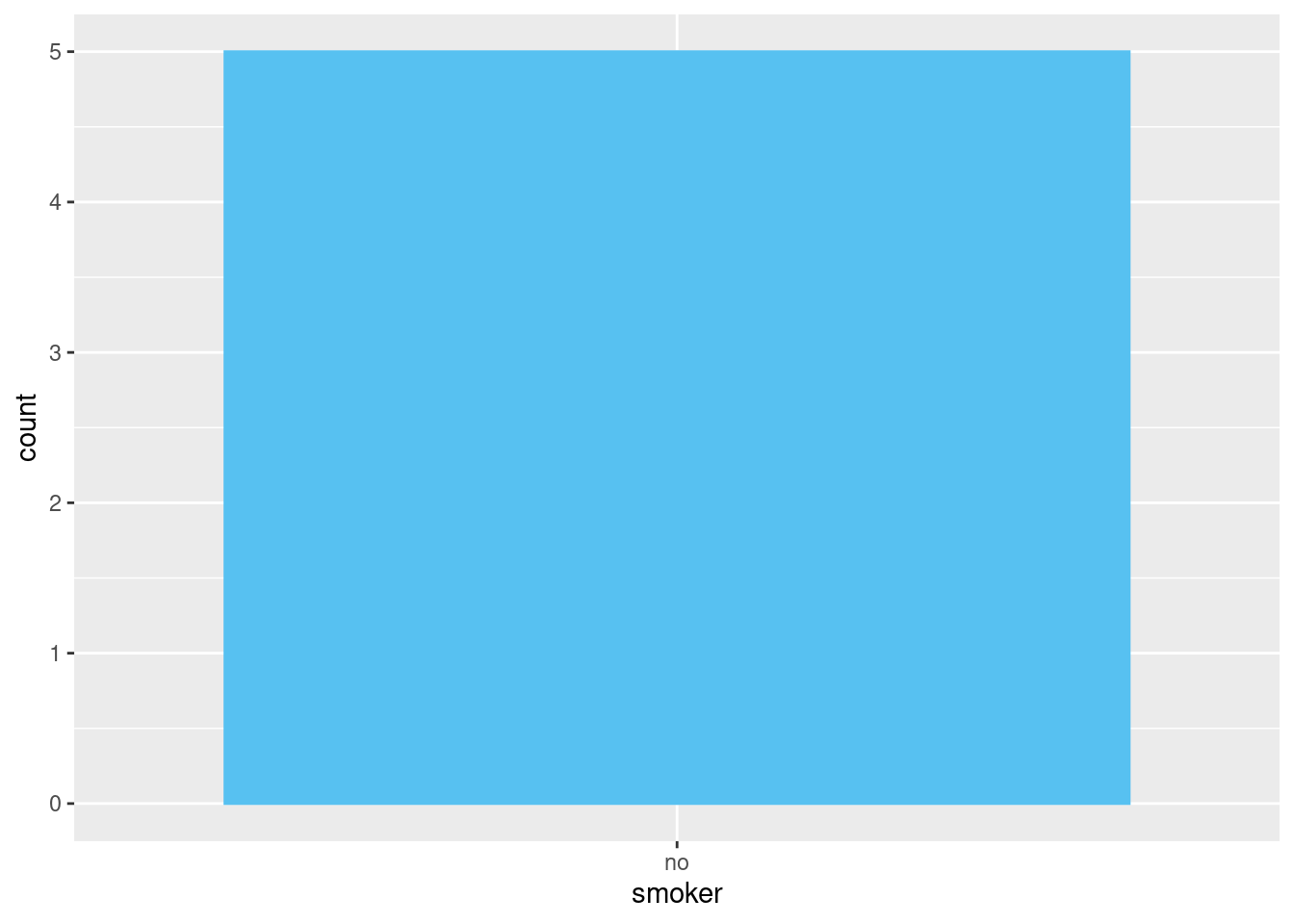
ggplot2::ggplot(
data = health,
mapping = ggplot2::aes(
x = .data[["smoker"]])
) +
ggplot2::geom_bar() +
ggplot2::scale_x_discrete(drop = FALSE)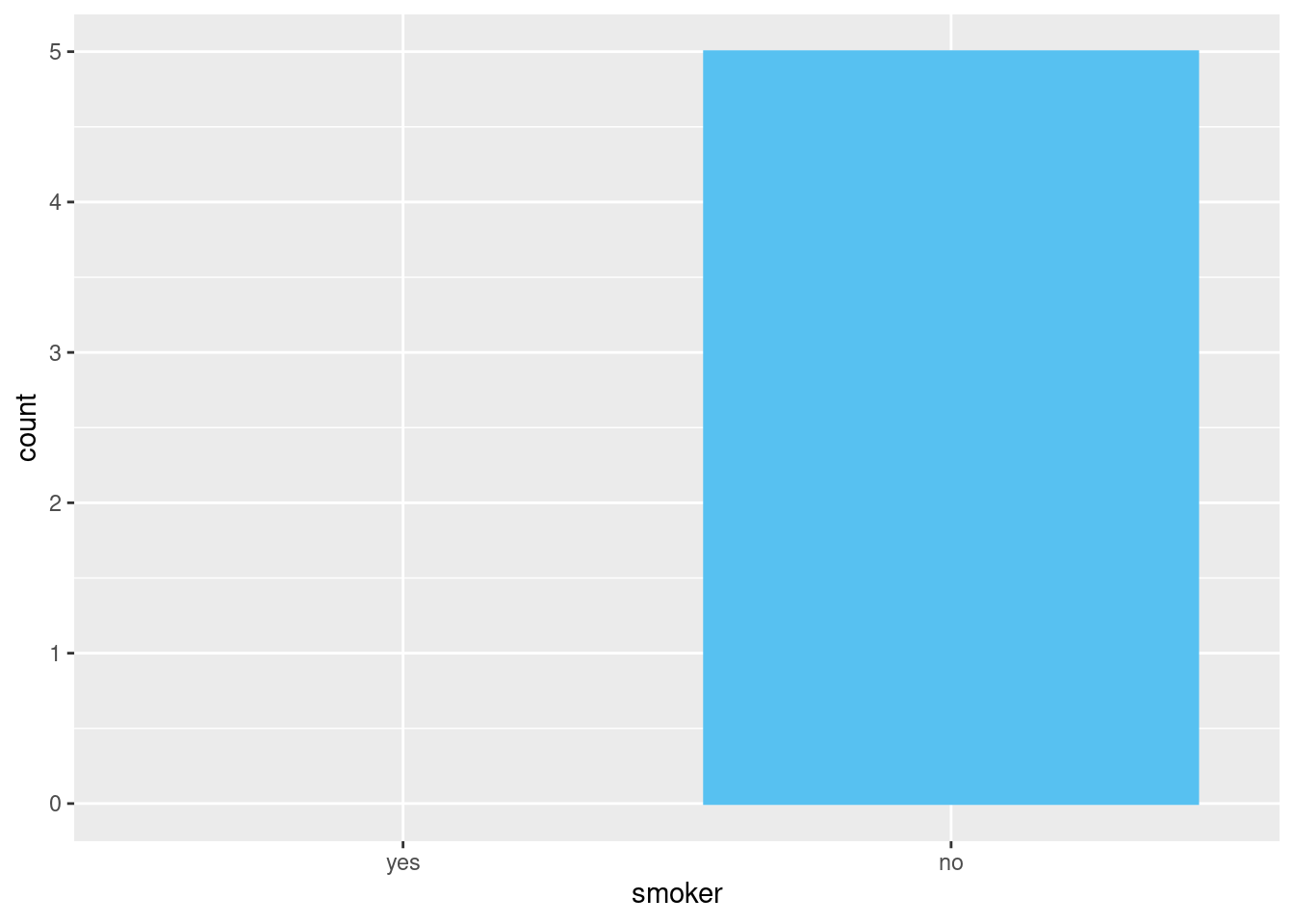
The same problem comes up more generally with dplyr::group_by(). And again you can use .drop = FALSE to preserve all factor levels:
health |>
dplyr::group_by(
.data[["smoker"]]
) |>
dplyr::summarize(
n = dplyr::n(),
mean_age = mean(.data[["age"]]),
min_age = min(.data[["age"]]),
max_age = max(.data[["age"]]),
sd_age = sd(.data[["age"]])
)## # A tibble: 1 × 6
## smoker n mean_age min_age max_age sd_age
## <fct> <int> <dbl> <dbl> <dbl> <dbl>
## 1 no 5 60 34 88 21.6health |>
dplyr::group_by(
.data[["smoker"]],
.drop = FALSE) |>
dplyr::summarize(
n = dplyr::n(),
mean_age = mean(.data[["age"]]),
min_age = min(.data[["age"]]),
max_age = max(.data[["age"]]),
sd_age = sd(.data[["age"]])
)## Warning: There were 2 warnings in `dplyr::summarize()`.
## The first warning was:
## ℹ In argument: `min_age = min(.data[["age"]])`.
## ℹ In group 1: `smoker = yes`.
## Caused by warning in `min()`:
## ! no non-missing arguments to min; returning Inf
## ℹ Run `dplyr::last_dplyr_warnings()` to see the 1 remaining warning.## # A tibble: 2 × 6
## smoker n mean_age min_age max_age sd_age
## <fct> <int> <dbl> <dbl> <dbl> <dbl>
## 1 yes 0 NaN Inf -Inf NA
## 2 no 5 60 34 88 21.6We get some interesting results here because when summarizing an empty group, the summary functions are applied to zero-length vectors
Here we see mean({zero_vec}) returning NaN because
mean({zero_vec}) = sum({zero_vec})/length({zero_vec})
which is 0/0.
max() and min() return -Inf and Inf for empty vectors.
health |>
dplyr::group_by(
.data[["smoker"]],
.drop = FALSE) |>
dplyr::summarize(
n = dplyr::n(),
mean_age = mean(.data[["age"]]),
min_age = min(.data[["age"]]),
max_age = max(.data[["age"]]),
sd_age = sd(.data[["age"]])
)## Warning: There were 2 warnings in `dplyr::summarize()`.
## The first warning was:
## ℹ In argument: `min_age = min(.data[["age"]])`.
## ℹ In group 1: `smoker = yes`.
## Caused by warning in `min()`:
## ! no non-missing arguments to min; returning Inf
## ℹ Run `dplyr::last_dplyr_warnings()` to see the 1 remaining warning.## # A tibble: 2 × 6
## smoker n mean_age min_age max_age sd_age
## <fct> <int> <dbl> <dbl> <dbl> <dbl>
## 1 yes 0 NaN Inf -Inf NA
## 2 no 5 60 34 88 21.6Instead of .drop = FALSE, we can use tidyr::complete() to the implicit missing values explicit. The main drawback of this approach is that you get an NA for the count, even though you know that it should be zero.
health |>
dplyr::group_by(
.data[["smoker"]]
) |>
dplyr::summarize(
n = dplyr::n(),
mean_age = mean(.data[["age"]]),
min_age = min(.data[["age"]]),
max_age = max(.data[["age"]]),
sd_age = sd(.data[["age"]])
) |>
tidyr::complete(.data[["smoker"]])## # A tibble: 2 × 6
## smoker n mean_age min_age max_age sd_age
## <fct> <int> <dbl> <dbl> <dbl> <dbl>
## 1 yes NA NA NA NA NA
## 2 no 5 60 34 88 21.618.4.2 forcats 1.0.0 Extra
Adapted from forcats 1.0.0 blog
There are two ways to represent a missing value in a factor:
NA as values:
## [1] "x" "y"NA as factors:
## [1] "x" "y" NAThey provide different behaviour when is.na and as.integer are applied
NA as values:
NAs in the values tend to be best for data analysis.
## [1] FALSE FALSE TRUE TRUE FALSE## [1] 1 2 NA NA 1NA as factors:
NAs in the levels are useful if you need to control where missing values are shown in a table or a plot
## [1] FALSE FALSE FALSE FALSE FALSE## [1] 1 2 3 3 1To make it easier to switch between these forms, forcats now comes fct_na_value_to_level() and fct_na_level_to_value().
In the plot below, we use fct_infreq() to reorder the levels of the factor so that the highest frequency levels are at the top of the bar chart. However, because the NAs are stored in the values, fct_infreq() has no ability to affect them, so they appear in their “default” position.
example <- data.frame(
hair_color = c(dplyr::starwars$hair_color,
rep("missing", 10),
rep("don't know", 5))
) |>
dplyr::mutate(
hair_color = .data[["hair_color"]] |>
# Reorder factor by frequency
forcats::fct_infreq() |>
# Group hair colours with less than 2 observations as Other
forcats::fct_lump_min(2, other_level = "(Other)") |>
forcats::fct_rev()
)
example |>
ggplot2::ggplot(
mapping = ggplot2::aes(
y = .data[["hair_color"]]
)
) +
ggplot2::geom_bar() +
ggplot2::labs(y = "Hair color")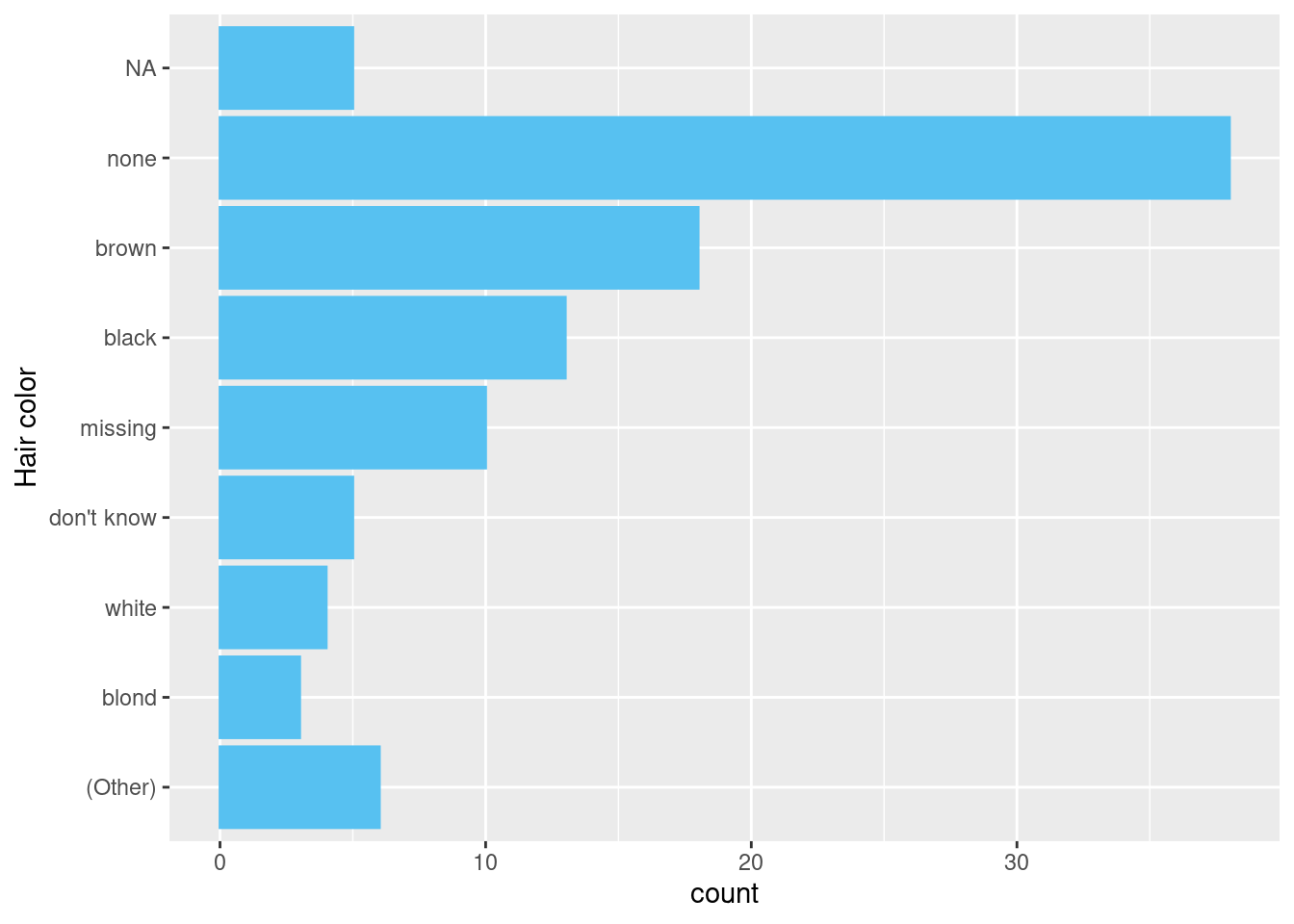
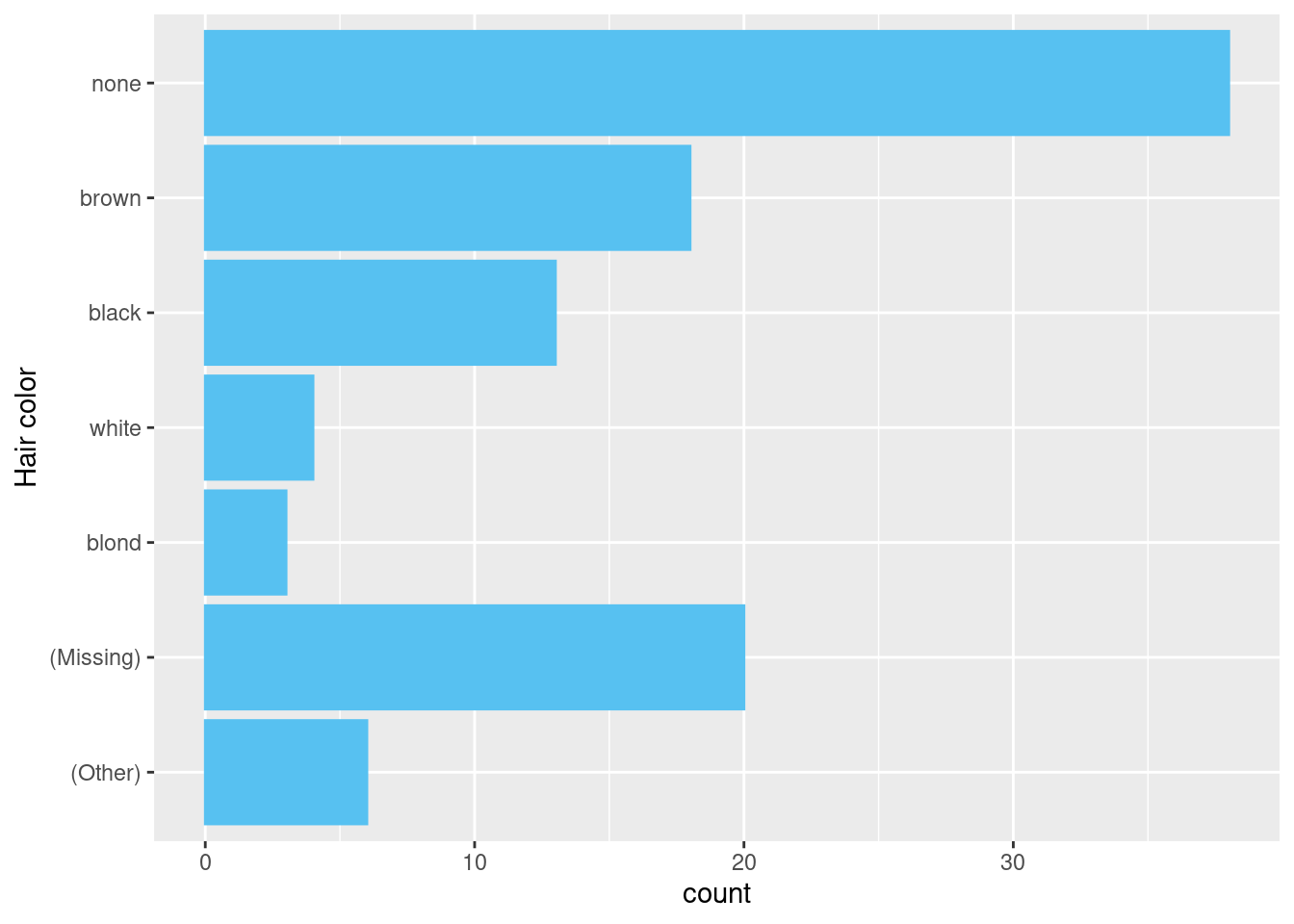
To consolidate all missing values,
Use
fct_recodeto convert “don’t know” to the value “missing”.Use
fct_na_level_to_value()to convert NA as a factor called “missing”.Use
fct_na_value_to_level()to convert NA to the value “missing”.
example <- data.frame(
hair_color = c(dplyr::starwars$hair_color,
rep("missing", 10),
rep("don't know", 5))
) |>
dplyr::mutate(
hair_color = .data[["hair_color"]] |>
# Reorder factor by frequency
forcats::fct_infreq() |>
forcats::fct_recode(
missing = "don't know") |>
forcats::fct_na_level_to_value(
extra_levels = "missing") |>
forcats::fct_na_value_to_level(
level = "(Missing)") |>
# Group hair colours with less than 2 observations as Other
forcats::fct_lump_min(2, other_level = "(Other)") |>
forcats::fct_rev()
)
example |>
ggplot2::ggplot(
mapping = ggplot2::aes(
y = .data[["hair_color"]]
)
) +
ggplot2::geom_bar() +
ggplot2::labs(y = "Hair color")