Exercises
1. How many rows are in penguins? How many columns?
## # A tibble: 344 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
## <fct> <fct> <dbl> <dbl> <int> <int>
## 1 Adelie Torgersen 39.1 18.7 181 3750
## 2 Adelie Torgersen 39.5 17.4 186 3800
## 3 Adelie Torgersen 40.3 18 195 3250
## 4 Adelie Torgersen NA NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193 3450
## 6 Adelie Torgersen 39.3 20.6 190 3650
## 7 Adelie Torgersen 38.9 17.8 181 3625
## 8 Adelie Torgersen 39.2 19.6 195 4675
## 9 Adelie Torgersen 34.1 18.1 193 3475
## 10 Adelie Torgersen 42 20.2 190 4250
## # ℹ 334 more rows
## # ℹ 2 more variables: sex <fct>, year <int>See first line. 344 rows x 8 columns.
2. What does the bill_depth_mm variable in the penguins data frame describe? Read the help for ?penguins to find out.
Call ?penguins for definition from package documentation
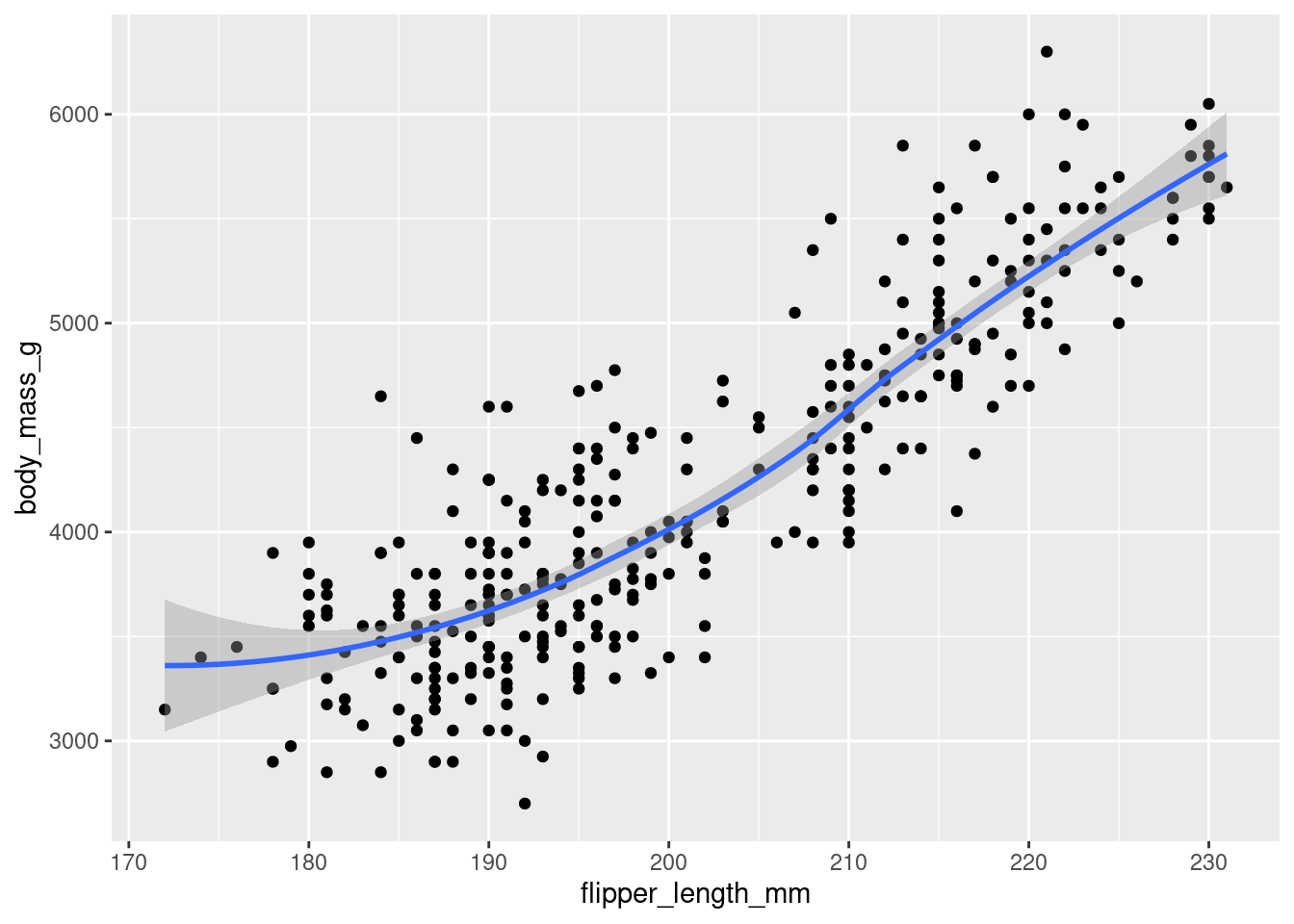
3. Make a scatterplot of bill_depth_mm vs. bill_length_mm. That is, make a scatterplot with bill_depth_mm on the y-axis and bill_length_mm on the x-axis. Describe the relationship between these two variables.

Positive, linear relationship? We’ll see more about this in later exercises.
4. What happens if you make a scatterplot of species vs. bill_depth_mm? What might be a better choice of geom?
## Warning: Removed 2 rows containing missing values or values outside the scale range
## (`geom_point()`).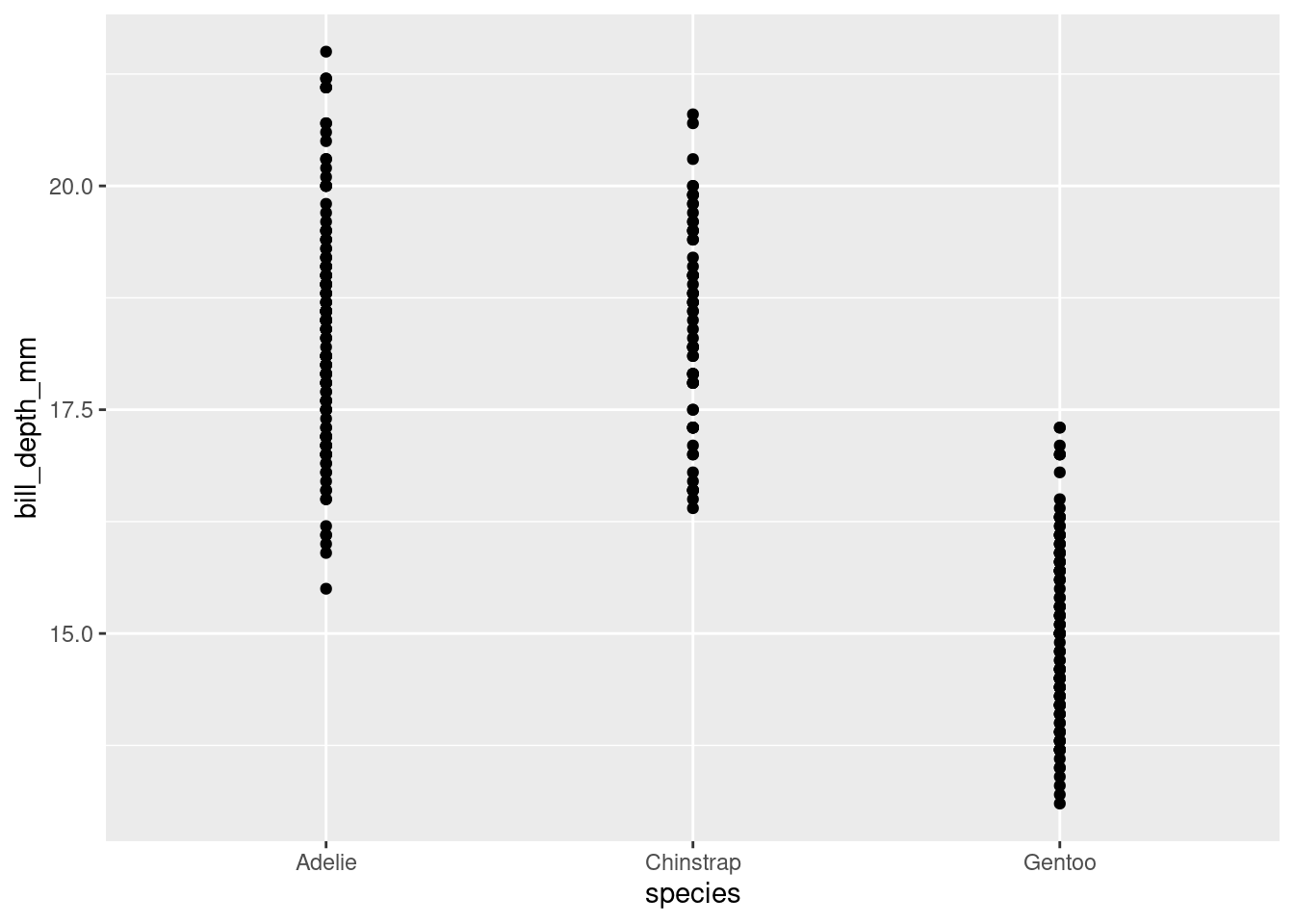
Dotplot for every species. Boxplot would tell us more.
## Warning: Removed 2 rows containing non-finite outside the scale range
## (`stat_boxplot()`).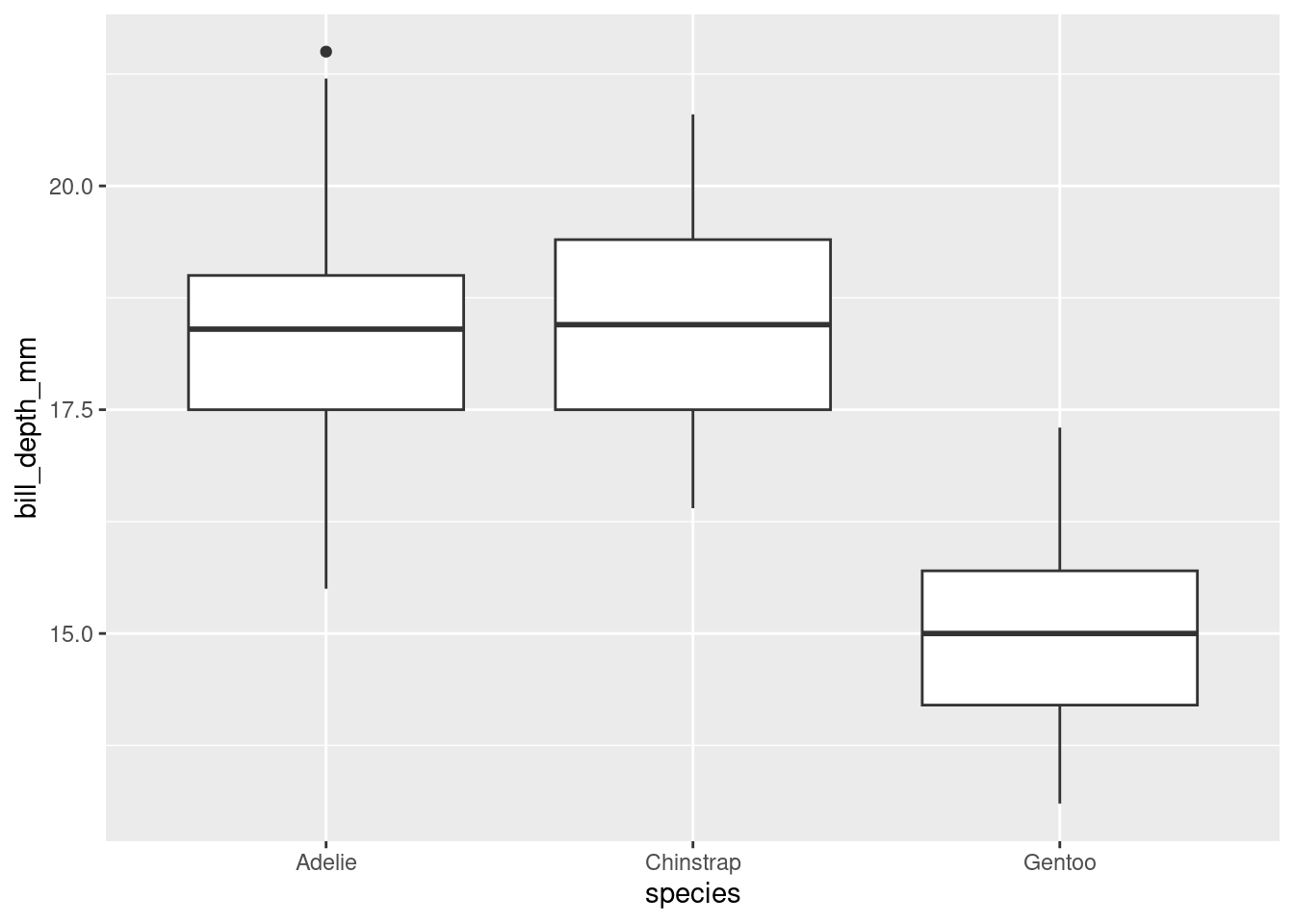
This better compares bill depths across species.
5. Why does the following give an error and how would you fix it?
The error reads ! geom_point() requires the following missing aesthetics: x and y. Call needs x variable and y variable.
6. What does the na.rm argument do in geom_point()? What is the default value of the argument? Create a scatterplot where you successfully use this argument set to TRUE.
From function documentation, “If FALSE, the default, missing values are removed with a warning. If TRUE, missing values are silently removed.”

7. Add the following caption to the plot you made in the previous exercise: “Data come from the palmerpenguins package.” Hint: Take a look at the documentation for labs().
caption argument adds a caption.
ggplot(penguins) +
geom_boxplot(aes(x = species, y = bill_depth_mm)) +
labs(
title = "Distribution of Penguin Bill Depths by Species",
subtitle = "For penguins at Palmer Station Antarctica",
x = "Species",
y = "Bill depth (in millimeters)",
caption = "Data come from the `palmerpenguins` package."
)
8.Recreate the following visualization. What aesthetic should bill_depth_mm be mapped to? And should it be mapped at the global level or at the geom level?
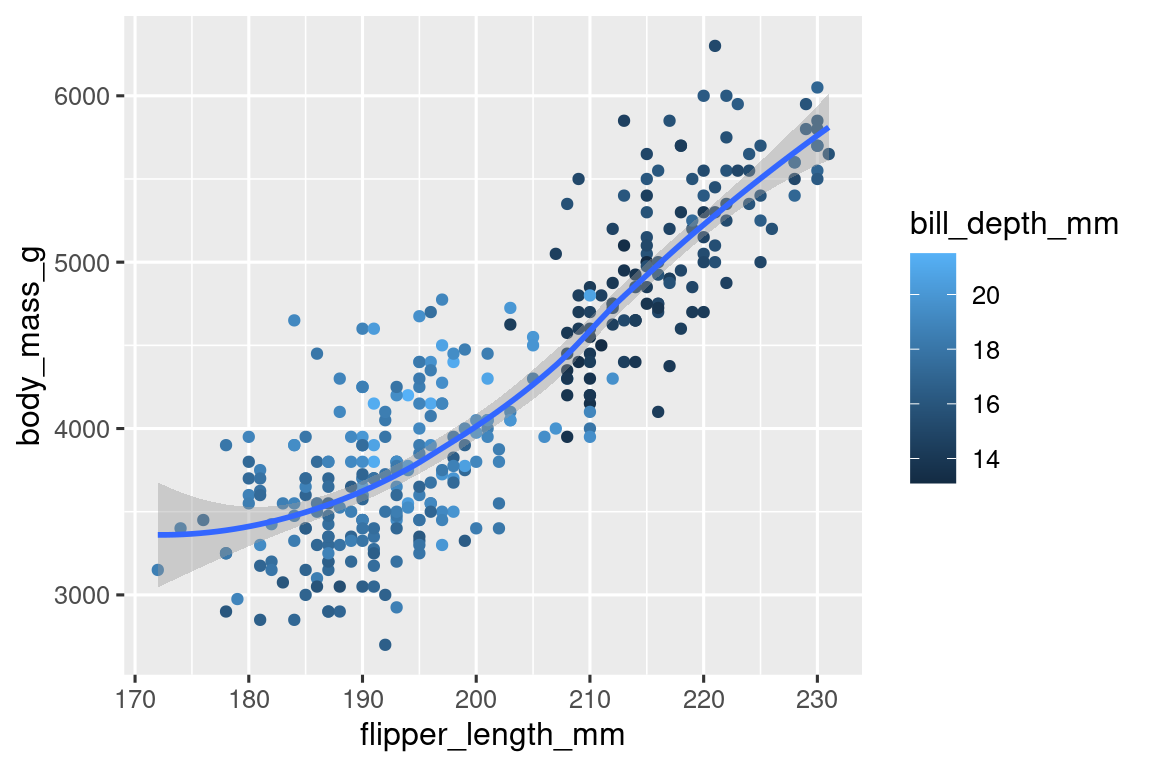
ggplot(data = penguins, aes(x = flipper_length_mm, y = body_mass_g)) +
geom_point(aes(color = bill_depth_mm)) +
geom_smooth(method = "loess")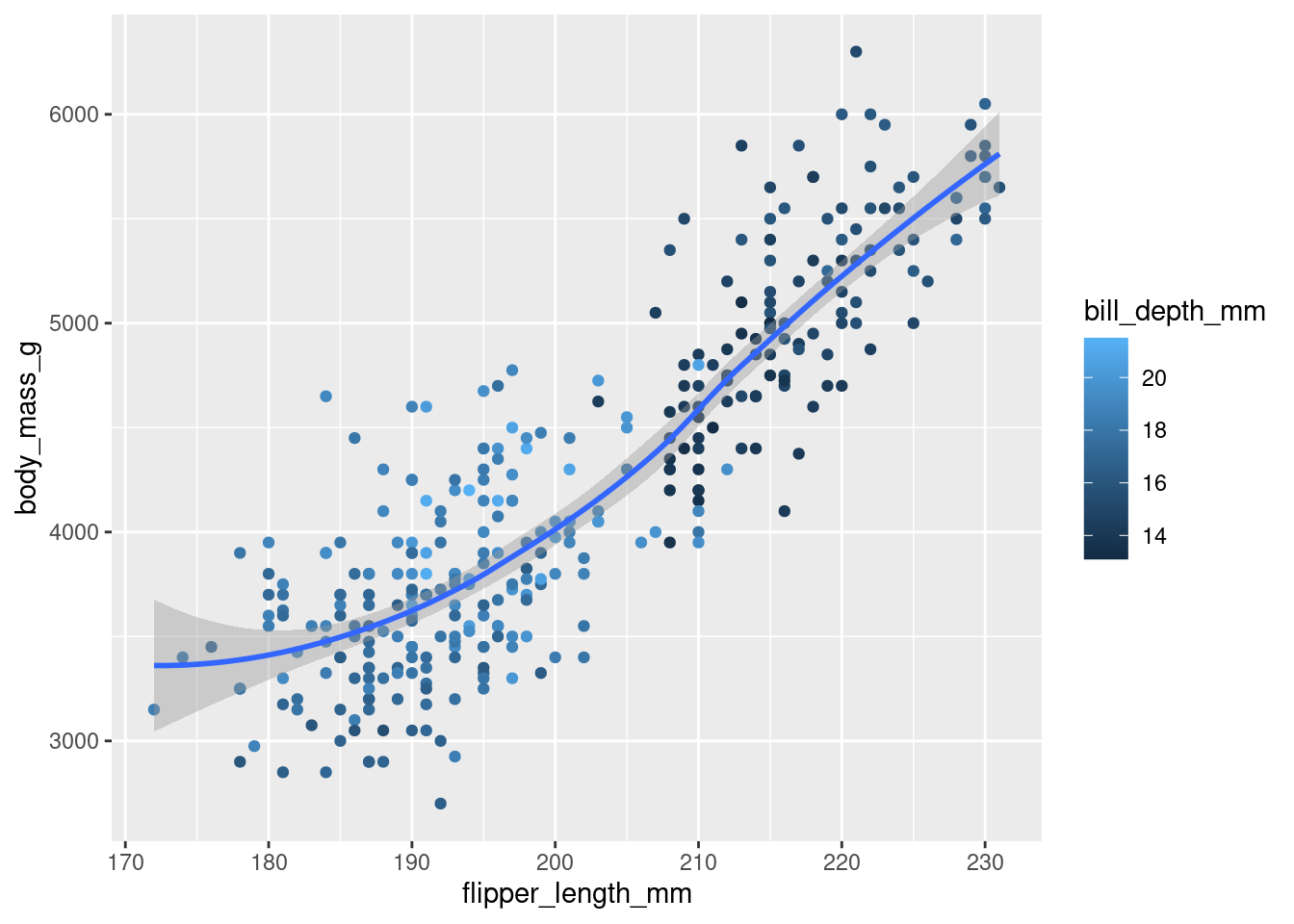
9. Run this code in your head and predict what the output will look like. Then, run the code in R and check your predictions.
ggplot(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g, color = island)
) +
geom_point() +
geom_smooth(se = FALSE)
10. Will these two graphs look different? Why/why not?
ggplot(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
) +
geom_point() +
geom_smooth()
ggplot() +
geom_point(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
) +
geom_smooth(
data = penguins,
mapping = aes(x = flipper_length_mm, y = body_mass_g)
)Only difference is mapping globally vs. locally, and mapped same in both geoms, so they should look the same.