15.4 NMDS
NMDS (Non-metric multidimensional scaling) is an iterative procedure trying to make the ordinated space more similar to the input matrix in each step.
# presence-absence matrix
pa = vegan::decostand(comm, "pa") # 100 rows (sites), 69 columns (species)
# keep only sites in which at least one species was found
pa = pa[rowSums(pa) != 0, ] # 84 rows, 69 columnsThe resulting matrix serves as input for the NMDS. k specifies the number of output axes, here, set to 4.
set.seed(25072018)
nmds = vegan::metaMDS(comm = pa, k = 4, try = 500)## Run 0 stress 0.08889093
## Run 1 stress 0.08911386
## ... Procrustes: rmse 0.01189629 max resid 0.05668371
## Run 2 stress 0.08889196
## ... Procrustes: rmse 0.01294593 max resid 0.08190055
## Run 3 stress 0.08839788
## ... New best solution
## ... Procrustes: rmse 0.01803938 max resid 0.07935802
## Run 4 stress 0.08874618
## ... Procrustes: rmse 0.01234385 max resid 0.06113602
## Run 5 stress 0.08906707
## Run 6 stress 0.08887794
## ... Procrustes: rmse 0.01717898 max resid 0.0620984
## Run 7 stress 0.08860411
## ... Procrustes: rmse 0.01095475 max resid 0.05457201
## Run 8 stress 0.091256
## Run 9 stress 0.08962818
## Run 10 stress 0.09265916
## Run 11 stress 0.09228074
## Run 12 stress 0.0901847
## Run 13 stress 0.09096006
## Run 14 stress 0.08844062
## ... Procrustes: rmse 0.005672234 max resid 0.03209942
## Run 15 stress 0.0887541
## ... Procrustes: rmse 0.01315372 max resid 0.06719546
## Run 16 stress 0.08832784
## ... New best solution
## ... Procrustes: rmse 0.007283695 max resid 0.03435924
## Run 17 stress 0.09083674
## Run 18 stress 0.09198497
## Run 19 stress 0.09025482
## Run 20 stress 0.0896162
## *** Best solution was not repeated -- monoMDS stopping criteria:
## 20: no. of iterations >= maxitnmds$stress## [1] 0.08832784A stress value of 9 [???] represents a very good result, which means that the reduced ordination space represents the large majority of the variance of the input matrix.
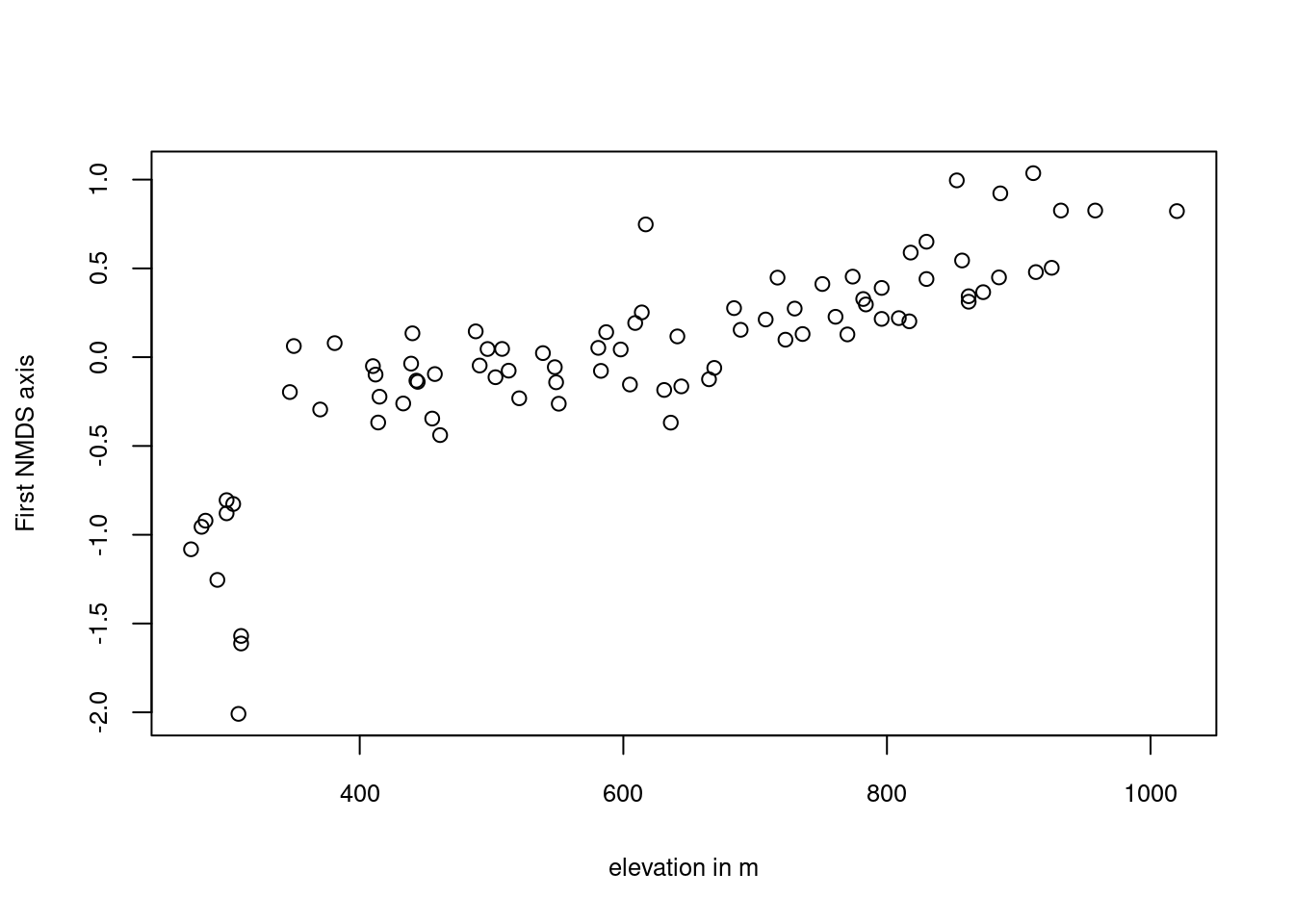
Since humidity is highly correlated with elevation, we rotate the NMDS axes in accordance with elevation.
elev = dplyr::filter(random_points, id %in% rownames(pa)) |>
dplyr::pull(dem)
# rotating NMDS in accordance with altitude (proxy for humidity)
rotnmds = vegan::MDSrotate(nmds, elev)
# extracting the first two axes
sc = vegan::scores(rotnmds, choices = 1:2, display = "sites")
# plotting the first axis against altitude
plot(y = sc[, 1], x = elev, xlab = "elevation in m",
ylab = "First NMDS axis", cex.lab = 0.8, cex.axis = 0.8)