15.5 Floristic Gradient
To predict the floristic gradient spatially, we use a random forest model (Tomislav Hengl et al. 2018). Random forest models are frequently applied in environmental and ecological modeling, and often provide the best results in terms of predictive performance (Schratz et al. 2019)
# construct response-predictor matrix
# id- and response variable
rp = data.frame(id = as.numeric(rownames(sc)), sc = sc[, 1])
# join the predictors (dem, ndvi and terrain attributes)
rp = inner_join(random_points, rp, by = "id")Decision trees split the predictor space into a number of regions.
tree_mo = tree::tree(sc ~ dem, data = rp)
plot(tree_mo)
text(tree_mo, pretty = 0)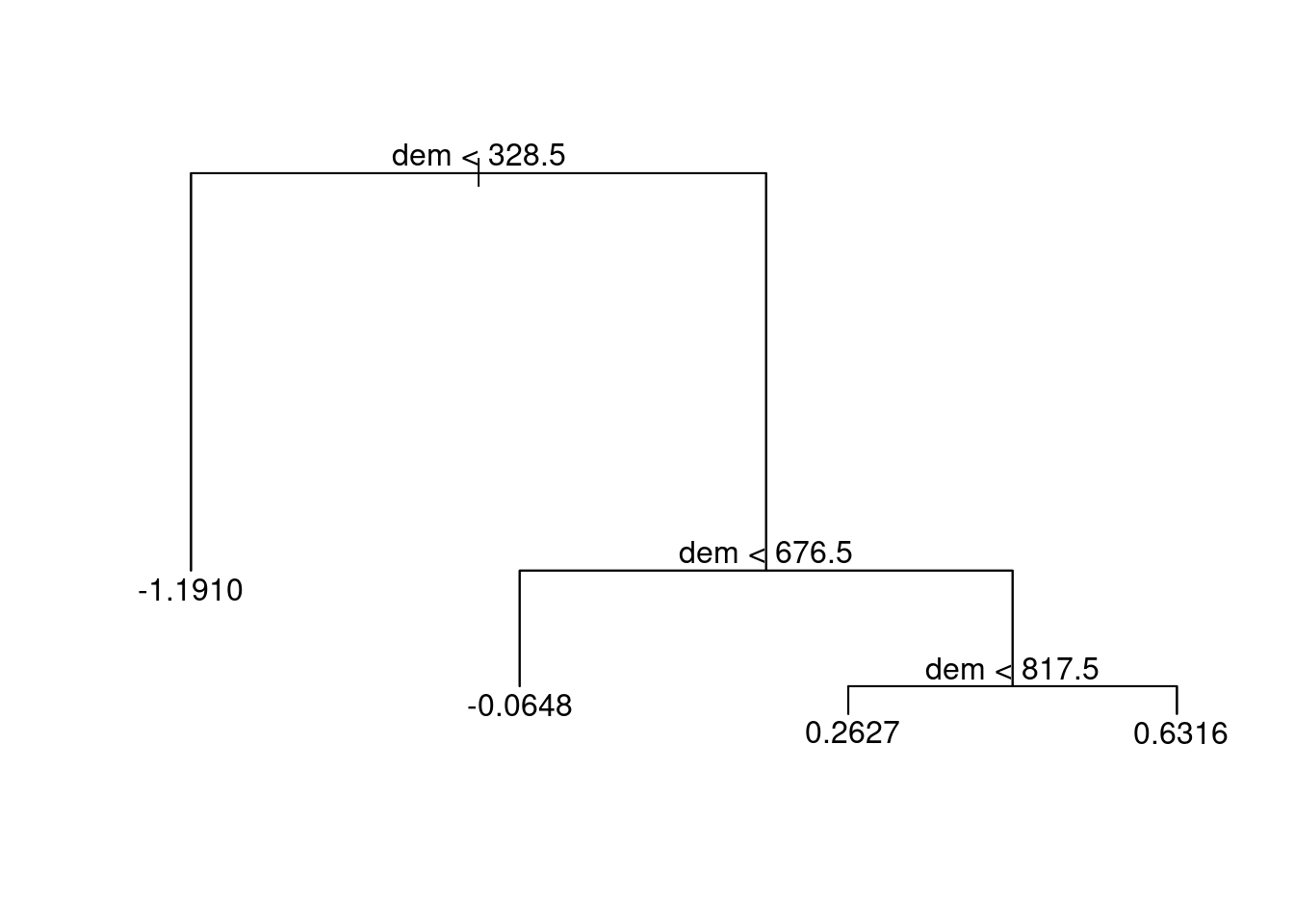 ## Tuning
## Tuning
- The response variable is numeric, hence a regression task will replace the classification task
- Instead of the AUROC which can only be used for categorical response variables, we will use the root mean squared error (RMSE) as performance measure
- We use a random forest model instead of a support vector machine which naturally goes along with different hyperparameters
Having already constructed the input variables (rp), we are all set for specifying the mlr3
# create task
task = mlr3spatiotempcv::as_task_regr_st(select(rp, -id, -spri),
id = "mongon", target = "sc")# one random forest with default hyperparameter settings
# e.g. proportion of observations, minimum node size
lrn_rf = lrn("regr.ranger", predict_type = "response")# specifying the search space
search_space = paradox::ps(
mtry = paradox::p_int(lower = 1, upper = ncol(task$data()) - 1),
sample.fraction = paradox::p_dbl(lower = 0.2, upper = 0.9),
min.node.size = paradox::p_int(lower = 1, upper = 10)
)# workflow
autotuner_rf = mlr3tuning::AutoTuner$new(
learner = lrn_rf,
resampling = mlr3::rsmp("spcv_coords", folds = 5), # spatial partitioning
measure = mlr3::msr("regr.rmse"), # performance measure
terminator = mlr3tuning::trm("evals", n_evals = 50), # specify 50 iterations
search_space = search_space, # predefined hyperparameter search space
tuner = mlr3tuning::tnr("random_search") # specify random search
)# hyperparameter tuning
set.seed(0412022)
autotuner_rf$train(task) #produces many outputsautotuner_rf$tuning_result## mtry sample.fraction min.node.size learner_param_vals x_domain regr.rmse
## 1: 4 0.8999753 7 <list[4]> <list[3]> 0.3763596