15.1 Data
data("study_area", "random_points", "comm", package = "spDataLarge")
dem = rast(system.file("raster/dem.tif", package = "spDataLarge"))
ndvi = rast(system.file("raster/ndvi.tif", package = "spDataLarge"))study_area## Simple feature collection with 1 feature and 1 field
## Geometry type: POLYGON
## Dimension: XY
## Bounding box: xmin: 795338.3 ymin: 8932349 xmax: 797283.2 ymax: 8934832
## Projected CRS: WGS 84 / UTM zone 17S
## # A tibble: 1 × 2
## name geometry
## * <chr> <POLYGON [m]>
## 1 study area ((795703.6 8934738, 795768.3 8934731, 795909.3 8934685, 796007.3 8…random_points## Simple feature collection with 100 features and 2 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: 795551.4 ymin: 8932370 xmax: 797242.3 ymax: 8934800
## Projected CRS: WGS 84 / UTM zone 17S
## First 10 features:
## id spri geometry
## 1 1 4 POINT (797178.6 8932755)
## 2 2 4 POINT (796749.3 8932621)
## 3 3 3 POINT (796815.7 8932739)
## 4 4 2 POINT (797023.3 8932600)
## 5 5 4 POINT (796647.3 8932692)
## 6 6 5 POINT (796428.7 8932474)
## 7 7 6 POINT (796909.1 8932892)
## 8 8 2 POINT (797242.3 8933006)
## 9 9 3 POINT (796278.5 8932370)
## 10 10 3 POINT (796651 8932429)# community matrix of visited sites and observed species
str(comm, give.attr = FALSE)## 'data.frame': 100 obs. of 69 variables:
## $ Alon_meri: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Alst_line: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Alte_hali: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Alte_porr: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Anth_eccr: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Apod_ferr: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Atri_rotu: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bacc_line: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bego_acut: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bego_spec: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bowl_palm: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Brom_spec: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Caes_spin: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Cala_alba: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Calc_pinn: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Chen_peti: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Chio_bent: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Cicl_laci: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Cist_ling: num 0.019 0 0.125 0 0.006 0.038 3 0 0.006 0 ...
## $ Cist_pani: num 0.5 0.5 0.5 0.05 0.5 0.006 15 0.063 0.028 0.063 ...
## $ Comm_fasc: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Cras_conn: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Crem_parv: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Crot_alni: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Cycl_math: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Cycl_spec: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Drym_cord: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Erod_spec: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Euph_viri: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Exod_pros: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Fuer_chil: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Fuer_lime: num 0.013 0.006 0 0 0.002 0 0 0 0 0 ...
## $ Haag_paca: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Hoff_pros: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Ipom_purp: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Ipom_spec: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Jalt_loma: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lant_scab: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lyci_lyci: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Ment_cord: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Nasa_uren: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Nico_pani: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Nico_spec: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Nola_humi: num 0.013 0.019 0 0 0 0.031 10 0 0 0.031 ...
## $ Ophr_peru: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Oxal_corn: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Oxal_mega: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Oxal_spec: num 0 0 0 0 0 0 0.063 0 0 0 ...
## $ Pala_spec: num 0 0 0 0 0 0 0 0.125 0.009 0.031 ...
## $ Pari_debi: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Pele_matu: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Pepe_spec: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Phil_peru: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Pros_pall: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Puya_ferr: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Puya_spec: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Quin_brev: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Salv_spec: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Sene_trux: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Sicy_bade: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Sola_mont: num 0 0 0.125 0 1 0.019 5 0 0 0 ...
## $ Sola_mult: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Sola_peru: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Sola_phyl: num 0 0.031 0 0.094 0 0.013 2 0 0 0 ...
## $ Sonc_olea: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Tigr_gran: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Till_purp: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Trop_pelt: num 0 0 0 0 0 0 0 0 0 0 ...
## $ unkn_Malv: num 0 0 0 0 0 0 0 0 0 0 ...dem #digital elevation model## class : SpatRaster
## dimensions : 117, 117, 1 (nrow, ncol, nlyr)
## resolution : 30.85, 30.85 (x, y)
## extent : 794599.1, 798208.6, 8931775, 8935384 (xmin, xmax, ymin, ymax)
## coord. ref. : WGS 84 / UTM zone 17S (EPSG:32717)
## source : dem.tif
## name : dem
## min value : 238
## max value : 1094ndvi #Normalized Difference Vegetation Index## class : SpatRaster
## dimensions : 117, 117, 1 (nrow, ncol, nlyr)
## resolution : 30.85, 30.85 (x, y)
## extent : 794599.1, 798208.6, 8931775, 8935384 (xmin, xmax, ymin, ymax)
## coord. ref. : WGS 84 / UTM zone 17S (EPSG:32717)
## source : ndvi.tif
## name : ndvi
## min value : -0.3983102
## max value : 0.3437578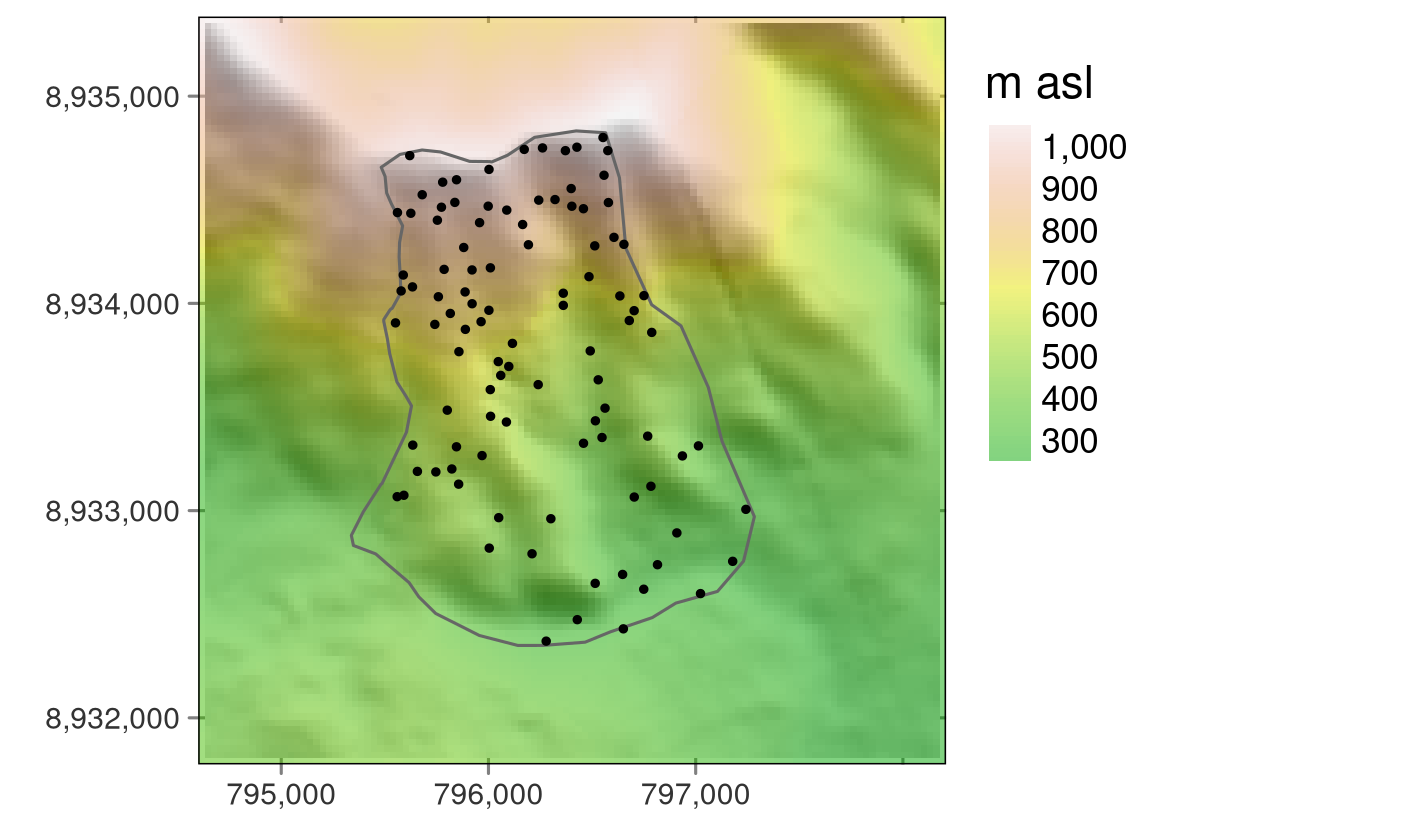
study area rasters