13.4 Tools for efficient grid search
A few tricks:

13.4.1 Submodel optimization
Types of models where, from a single model fit, multiple tuning parameters can be evaluated without refitting:
Partial Least Squares (no. of components to retain)
Boosting models (no. of boosting iterations, i.e. trees)
glmnetmakes (across the amount of regularization)MARSadds a set of nonlinear features (number of terms to retain)
The
tunepackage automatically applies this type of optimization whenever an applicable model is tuned. See also this vignette
methods("multi_predict")## [1] multi_predict._C5.0* multi_predict._earth*
## [3] multi_predict._elnet* multi_predict._glmnetfit*
## [5] multi_predict._lognet* multi_predict._multnet*
## [7] multi_predict._torch_mlp* multi_predict._train.kknn*
## [9] multi_predict._xgb.Booster* multi_predict.default*
## see '?methods' for accessing help and source codeparsnip:::multi_predict._C5.0 %>%
formals() %>%
names()## [1] "object" "new_data" "type" "trees" "..."For example, if a C5.0 model is fit to this cell classification data challenge, we can tune the trees. With all other parameters set at their default values, we can rapidly evaluate iterations from 1 to 100 :
data(cells)
cells <- cells %>% select(-case)
cell_folds <- vfold_cv(cells)
roc_res <- metric_set(roc_auc)
c5_spec <-
boost_tree(trees = tune()) %>%
set_engine("C5.0") %>%
set_mode("classification")
set.seed(2)
c5_tune <- c5_spec %>%
tune_grid(
class ~ .,
resamples = cell_folds,
grid = data.frame(trees = 1:100),
metrics = roc_res
)Even though we fit the model without the submodel prediction trick, this optimization is automatically applied by
parsnip.
autoplot(c5_tune)
ggsave("images/13_c5_submodel.png")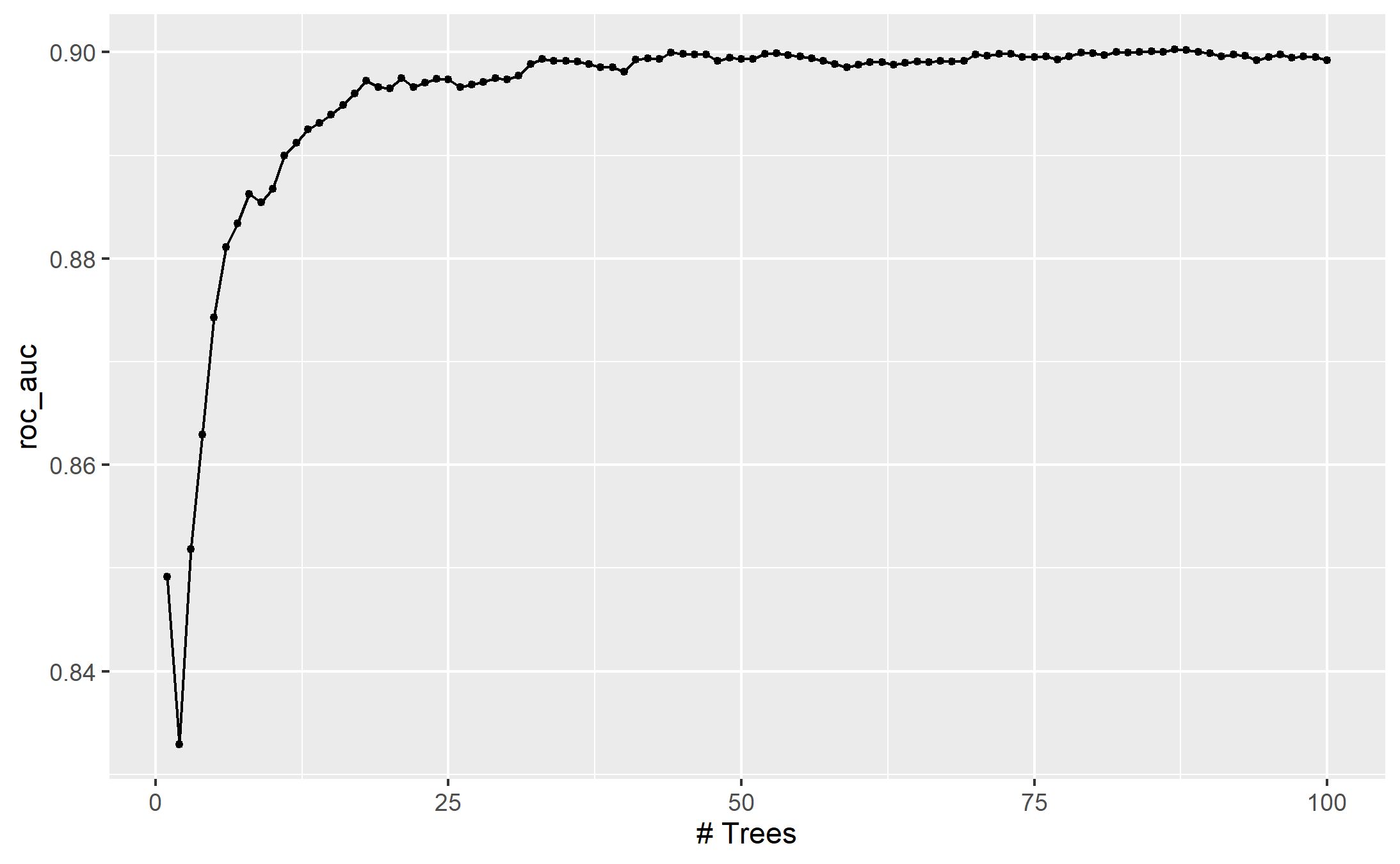
13.4.2 Parallel processing
backend packages right now are doFuture, doMC, doMPI, doParallel, doRedis,doRNG, doSNOW, and doAzureParallel
In tune_*(), there are two approaches, often set in control_grid() or control_resamples()
parallel_over = "resamplesorparallel_over = "everything"orparallel_over = NULL(the default) chooses “resamples” if there are more than one resample, otherwise chooses “everything” to attempt to maximize core utilization
Note that switching between parallel_over strategies is not guaranteed to use the same random number generation schemes. However, re-tuning a model using the same parallel_over strategy is guaranteed to be reproducible between runs.
On a shared server, never never consume all of the cores.
all_cores <- parallel::detectCores(logical = FALSE)
library(doParallel)
cl <- makePSOCKcluster(all_cores)
doParallel::registerDoParallel(cl)Be careful to avoid use of variables from the global environment. For example:
num_pcs <- 3
recipe(mpg ~ ., data = mtcars) %>%
# Bad since num_pcs might not be found by a worker process
step_pca(all_predictors(), num_comp = num_pcs)
recipe(mpg ~ ., data = mtcars) %>%
# Good since the value is injected into the object
step_pca(all_predictors(), num_comp = !!num_pcs)for the most part, the logging provided by tune_grid() will not be seen when running in parallel.
13.4.3 Benchmarking Parallel with boosted trees
Three scenarios
Preprocess the data prior to modeling using
dplyrConduct the same preprocessing via a
recipeWith a
recipe, add a step that has a high computational cost
using variable numbers of worker processes and using the two parallel_over options, on a computer with 10 physical cores
For dplyr and the simple recipe
There is little difference in the execution times between the panels.
There is some benefit for using
parallel_over = "everything"with many cores. However, as shown in the figure, the majority of the benefit of parallel processing occurs in the first five workers.
With the expensive preprocessing step, there is a considerable difference in execution times. Using parallel_over = "everything" is problematic since, even using all cores, it never achieves the execution time that parallel_over = "resamples" attains with just five cores. This is because the costly preprocessing step is unnecessarily repeated in the computational scheme.
Overall, note that the increased computational savings will vary from model-to-model and are also affected by the size of the grid, the number of resamples, etc. A very computationally efficient model may not benefit as much from parallel processing.
13.4.4 Racing Methods
The finetune package contains functions for racing.
One issue with grid search is that all models need to be fit across all resamples before any tuning parameters can be evaluated. It would be helpful if instead, at some point during tuning, an interim analysis could be conducted to eliminate any truly awful parameter candidates.
In racing methods the tuning process evaluates all models on an initial subset of resamples. Based on their current performance metrics, some parameter sets are not considered in subsequent resamples.
As an example, in the Chicago multilayer perceptron tuning process with a regular grid above, what would the results look like after only the first three folds?
We can fit a model where the outcome is the resampled area under the ROC curve and the predictor is an indicator for the parameter combination. The model takes the resample-to-resample effect into account and produces point and interval estimates for each parameter setting. The results of the model are one-sided 95% confidence intervals that measure the loss of the ROC value relative to the currently best performing parameters.
Any parameter set whose confidence interval includes zero would lack evidence that its performance is not statistically different from the best results. We retain 10 settings; these are resampled more. The remaining 10 submodels are no longer considered.
Racing methods can be more efficient than basic grid search as long as the interim analysis is fast and some parameter settings have poor performance. It also is most helpful when the model does not have the ability to exploit submodel predictions.
The tune_race_anova() function conducts an Analysis of Variance (ANOVA) model to test for statistical significance of the different model configurations.
library(finetune)
set.seed(99)
mlp_sfd_race <-
mlp_wflow %>%
tune_race_anova(
Chicago_folds,
grid = 20,
param_info = mlp_param,
metrics = rmse_mape_rsq_iic,
control = control_race(verbose_elim = TRUE)
)
write_rds(mlp_sfd_race,
"data/13-Chicago-mlp_sfd_race.rds",
compress = "gz")autoplot(mlp_sfd_race)
ggsave("images/13_mlp_sfd_race.png",
width = 12)
show_best(mlp_sfd_race, n = 6) hidden_units penalty epochs num_comp .metric .estimator mean n
<int> <dbl> <int> <int> <chr> <chr> <dbl> <int>
1 6 3.08e- 5 126 3 rmse standard 2.47 8
2 8 2.15e- 1 148 9 rmse standard 2.48 8
3 10 9.52e- 3 157 3 rmse standard 2.55 8
4 6 2.60e-10 84 12 rmse standard 2.56 8
5 5 1.48e- 2 94 4 rmse standard 2.57 8
6 4 7.08e- 1 98 14 rmse standard 2.60 8
# ... with 2 more variables: std_err <dbl>, .config <chr>
Warning message:
No value of `metric` was given; metric 'rmse' will be used.