10.3 Resampling methods
This diagram from the Hands-on machine learning in R from Boehmke & Greenwell illustrates how resampling fits into the general modeling workflow:
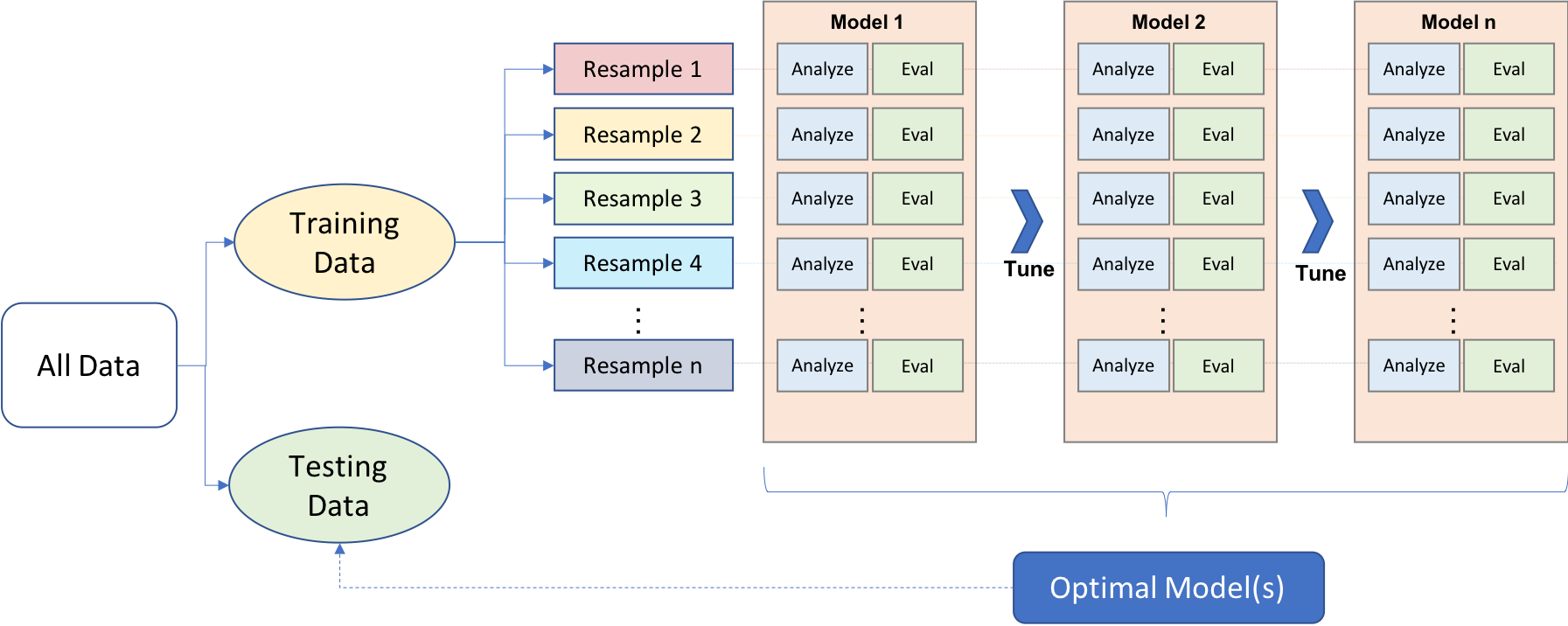
We can see that in resampling:
- happens after the data split into training and test sets
- some data is used for analyzing (analysis set), and some for evaluation (evaluation or assessment set, we’ll use assessment set from now on)
- this is an iterative as it can be repeated many times - it only applies to the training data
Effectively,
- the model is fit with the analysis set
- the model is evaluated with the evaluation set
The overall, final performance evaluation for the model is the average of the performance metrics of the n evaluation sets.
The way in which these analysis and evaluation sets are created define what the resampling techniques are specifically. We’ll go through the common ones:
cross-validation
repeated cross-validation
leave-one-out cross-validation
monte-carlo cross-validation
10.3.1 Cross-validation
Cross-validation is one of the most popular resampling techniques. There are several flavors
10.3.1.1 V-fold cross-validation
V-fold cross-validation involves the random splitting of the training data into approximately equal-sized “folds”. The diagram below illustrates v-fold cross validation with v set to 3 folds.
We can see that the training data points have
been randomly assigned to roughly equal-sized folds (in this case,
exactly equal) and that the assessment set for each fold is 2/3 of the
training data. Max and Julia note that while 3-fold CV is good to use
for illustrative purposed, it is not good in practice - in practice,
rather 5- or 10- fold CV is preferred.
set.seed(55)
ames_folds <- vfold_cv(ames_train, v = 10)
ames_folds## # 10-fold cross-validation
## # A tibble: 10 × 2
## splits id
## <list> <chr>
## 1 <split [2107/235]> Fold01
## 2 <split [2107/235]> Fold02
## 3 <split [2108/234]> Fold03
## 4 <split [2108/234]> Fold04
## 5 <split [2108/234]> Fold05
## 6 <split [2108/234]> Fold06
## 7 <split [2108/234]> Fold07
## 8 <split [2108/234]> Fold08
## 9 <split [2108/234]> Fold09
## 10 <split [2108/234]> Fold10The output contains info on how the training data was split: ~2000 are
in the analysis set, and ~220 are in the assessment set. You can
recuperate these sets by calling analysis() or assessment().
In chapter 5, we introduced the idea of stratified sampling, which is
almost always useful but particularly in cases with class imbalance. You
can perform V-fold CV with stratified samplying by using the strata
argument in the vfold_cv() call.
10.3.1.2 Repeated cross validation
V-fold CV introduced above may produce noisy estimates. A technique that averages over more than V statistics may be more appropriate to reduce the noise. This technique is repeated cross-validation, where we create R repeats of V-fold CV. Instead of averaging over V statistics, we are now averaging over V x R statistics:
vfold_cv(ames_train, v = 10, repeats = 5) ## # 10-fold cross-validation repeated 5 times
## # A tibble: 50 × 3
## splits id id2
## <list> <chr> <chr>
## 1 <split [2107/235]> Repeat1 Fold01
## 2 <split [2107/235]> Repeat1 Fold02
## 3 <split [2108/234]> Repeat1 Fold03
## 4 <split [2108/234]> Repeat1 Fold04
## 5 <split [2108/234]> Repeat1 Fold05
## 6 <split [2108/234]> Repeat1 Fold06
## 7 <split [2108/234]> Repeat1 Fold07
## 8 <split [2108/234]> Repeat1 Fold08
## 9 <split [2108/234]> Repeat1 Fold09
## 10 <split [2108/234]> Repeat1 Fold10
## # ℹ 40 more rows10.3.1.4 Monte Carlo cross validation (MCCV)
It’s like V-fold CV in the sense that training data is allocated to the assessment set with some fixed proportion. The difference is the resampling objects generated by MCCV are not mutually exclusive as the same data points can appear in the assessment set multiple times.
mc_cv(ames_train, prop = 9/10, times = 20)## # Monte Carlo cross-validation (0.9/0.1) with 20 resamples
## # A tibble: 20 × 2
## splits id
## <list> <chr>
## 1 <split [2107/235]> Resample01
## 2 <split [2107/235]> Resample02
## 3 <split [2107/235]> Resample03
## 4 <split [2107/235]> Resample04
## 5 <split [2107/235]> Resample05
## 6 <split [2107/235]> Resample06
## 7 <split [2107/235]> Resample07
## 8 <split [2107/235]> Resample08
## 9 <split [2107/235]> Resample09
## 10 <split [2107/235]> Resample10
## 11 <split [2107/235]> Resample11
## 12 <split [2107/235]> Resample12
## 13 <split [2107/235]> Resample13
## 14 <split [2107/235]> Resample14
## 15 <split [2107/235]> Resample15
## 16 <split [2107/235]> Resample16
## 17 <split [2107/235]> Resample17
## 18 <split [2107/235]> Resample18
## 19 <split [2107/235]> Resample19
## 20 <split [2107/235]> Resample2010.3.2 Validation sets
Another way you can assess the performance of your candidate model(s) - before moving forward to the test set - is to use a validation set. This might be an attractive option if you have big data. As the diagram below shows, the validation set is independent of the training data.
You can create your validation set by calling on validation_split()
and setting the proportion desired. There is also a strata argument to
conduct stratified sampling.
set.seed(12)
val_set <- validation_split(ames_train, prop = 3/4)
val_set## # Validation Set Split (0.75/0.25)
## # A tibble: 1 × 2
## splits id
## <list> <chr>
## 1 <split [1756/586]> validation10.3.3 Boostrapping
Bootstrapping can be used to estimate model performance. It’s good to be
aware that while it does produce lower variance compared to other
resampling methods, it has “significant pessimistic bias”. It also works
differently than other resampling methods. In the diagram below, we can
see that the analysis set is always equal to the size of the whole
training set, and we can also see that the same points can be selected
multiple times. The assessment sets contain the data points that were
not previously included in the analysis sets. Furthermore, these
assessment sets are not of the same size, as we’re about to see when we
call bootstraps().
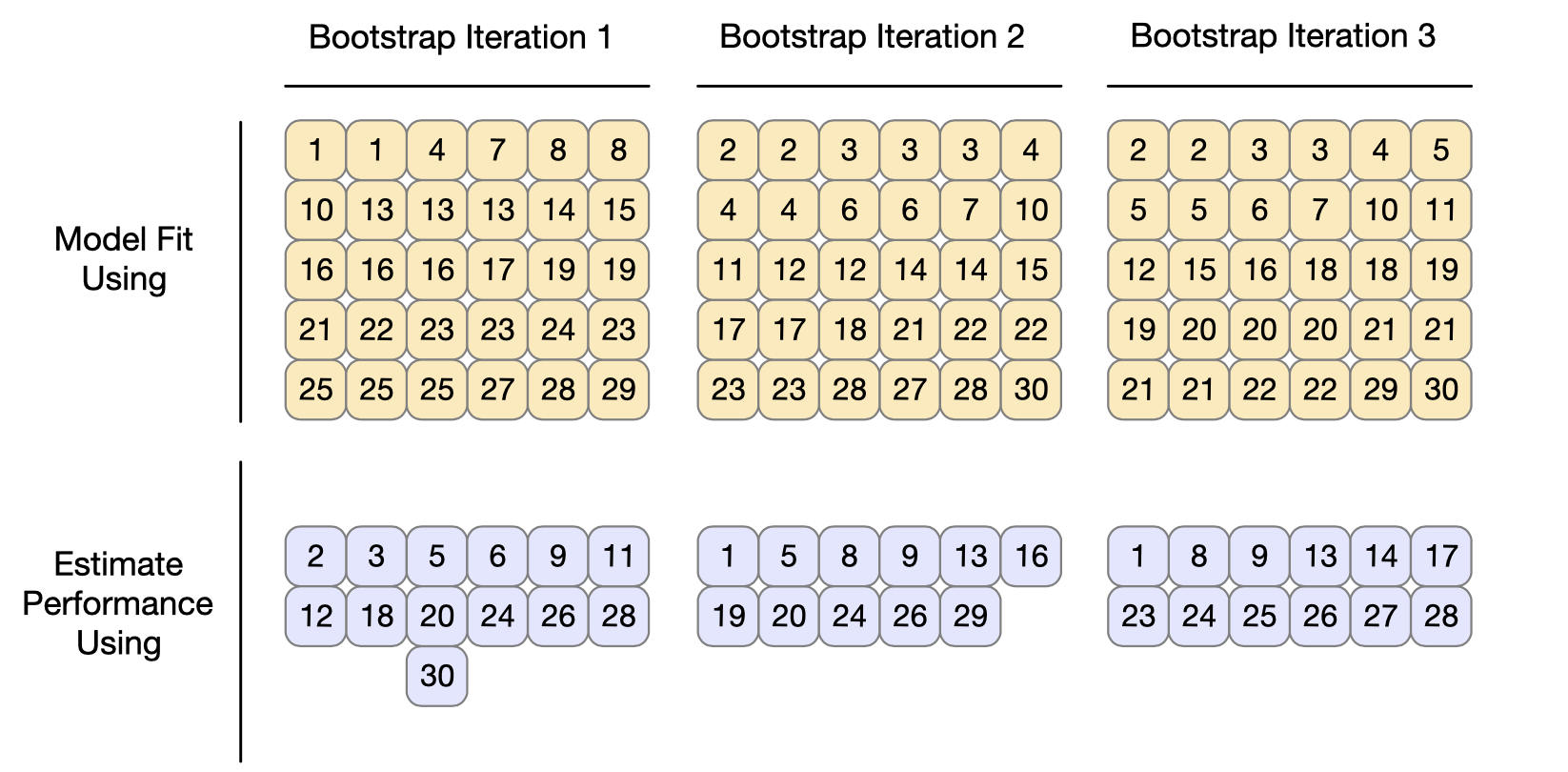 Operationally, performing
bootstrap resampling involves specifying the number of bootstrap samples
via the
Operationally, performing
bootstrap resampling involves specifying the number of bootstrap samples
via the times argument. There is also a strata argument for
conducting stratified sampling.
bootstraps(ames_train, times = 5)## # Bootstrap sampling
## # A tibble: 5 × 2
## splits id
## <list> <chr>
## 1 <split [2342/830]> Bootstrap1
## 2 <split [2342/867]> Bootstrap2
## 3 <split [2342/856]> Bootstrap3
## 4 <split [2342/849]> Bootstrap4
## 5 <split [2342/888]> Bootstrap510.3.4 Rolling forecasting origin resampling
Resampling with time series data needs a special setup as random sampling can ignore important trends such as seasonality. Rolling forecast resampling involves specifying the size of the analysis and assessment sets, and each iteration after the first one skips by a set number as the diagram illustrates below (with a skip of 1 as an example):
This time series resampling is done with rolling_origin. You can
specify the number of samples to be used for analysis with initial,
the number of samples used for each assessment resample with assess,
and cumulative set to true if you want the analysis resample to grow
beyong the size specified with initial. Additional arguments include
the skip and lag.
rolling_origin(data, initial = 5, assess = 1, cumulative = TRUE, skip = 0, lag = 0)