21.5 Inference with lower level helpers
Most useful for models not supported by anova, or parsnip, etc or when you want to have more control.
formulas <- list(
"full" = as.formula(avg_velocity ~ avg_elevation_gain + distance + aid_stations),
"partial" = as.formula(avg_velocity ~ avg_elevation_gain + distance),
"minimal" = as.formula(avg_velocity ~ avg_elevation_gain)
)
lm_spec <- linear_reg() %>% set_engine("lm")
AICs <- map(formulas, function(formula) {
lm_spec %>%
fit(formula, data = race_top_results) %>%
extract_fit_engine() %>%
AIC()
})
AICs## $full
## [1] 3123.333
##
## $partial
## [1] 3122.497
##
## $minimal
## [1] 3148.007How to determine whether these differences are significant? One possible solution is bootstrapping which can also be used when there are no nice theoretical properties.
velocity_model_summaries <- race_top_results %>%
bootstraps(times = 1000, apparent = TRUE) %>%
mutate(
full_aic = map_dbl(splits, ~ fit(lm_spec, formulas[["full"]], data = analysis(.x)) %>% extract_fit_engine() %>% AIC()),
partial_aic = map_dbl(splits, ~ fit(lm_spec, formulas[["partial"]], data = analysis(.x)) %>% extract_fit_engine() %>% AIC()),
minimal_aic = map_dbl(splits, ~ fit(lm_spec, formulas[["minimal"]], data = analysis(.x)) %>% extract_fit_engine() %>% AIC())
) %>%
select(full_aic, partial_aic, minimal_aic)velocity_model_summaries %>%
pivot_longer(c(full_aic, partial_aic, minimal_aic)) %>%
ggplot(aes(x = name, y = value)) +
geom_boxplot()
velocity_model_summaries %>%
summarize(
full_vs_partial = mean(full_aic < partial_aic),
partial_vs_minimal = mean(partial_aic < minimal_aic)
)## # A tibble: 1 × 2
## full_vs_partial partial_vs_minimal
## <dbl> <dbl>
## 1 0.377 0.916velocity_model_summaries %>%
unnest(full_coeffs) %>%
ggplot(aes(x = estimate)) +
geom_histogram(bins = 5) +
facet_wrap(~term, scales = "free_x") +
geom_vline(xintercept = 0, col = "green")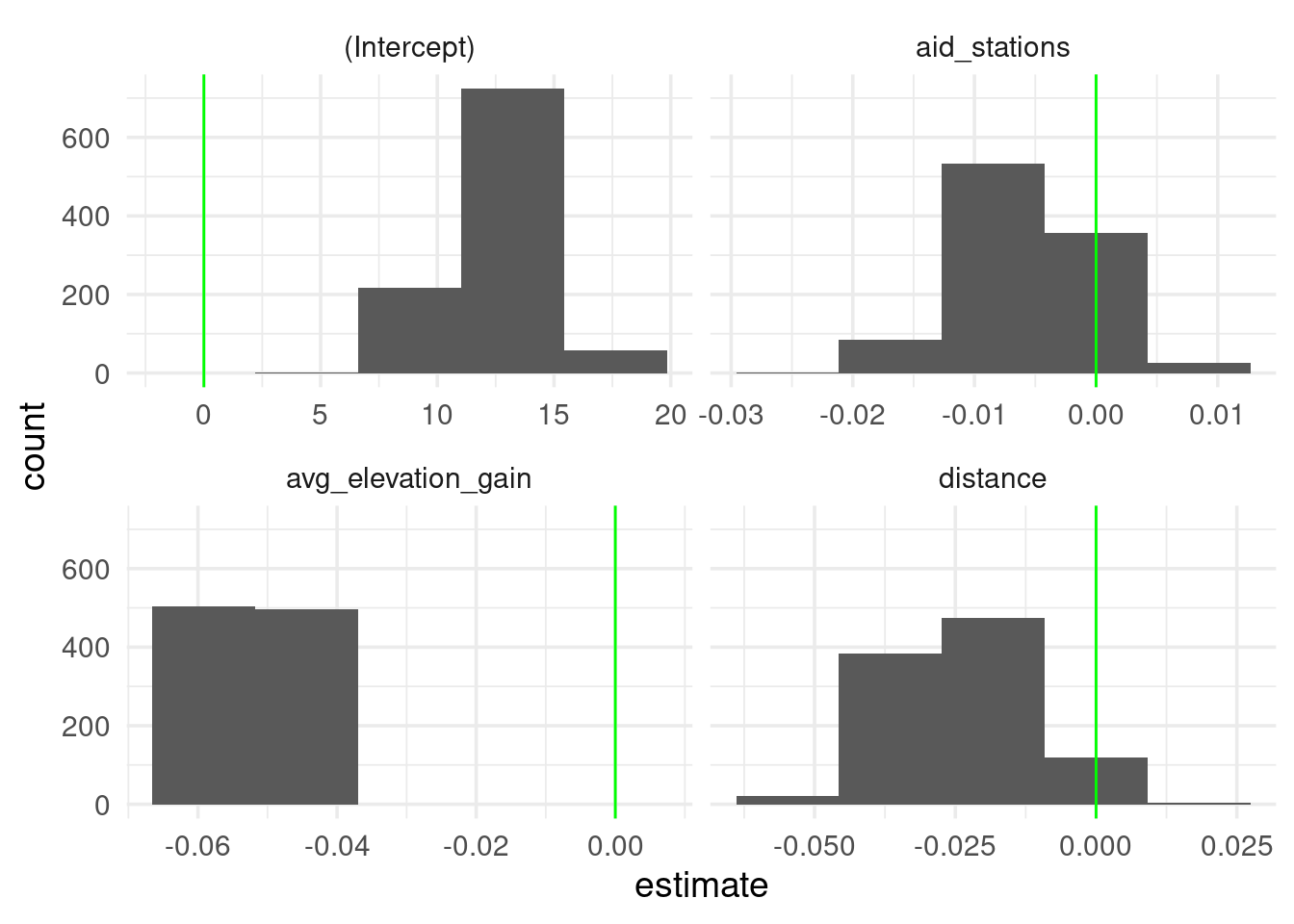
Small reps used for speed
race_top_results %>%
bootstraps(times = 20) %>%
mutate(
full_coeffs = map(splits, ~ fit(lm_spec, formulas[["full"]], data = analysis(.x)) %>% tidy())
) %>%
int_pctl(full_coeffs)## # A tibble: 4 × 6
## term .lower .estimate .upper .alpha .method
## <chr> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 (Intercept) 9.89 12.9 15.5 0.05 percentile
## 2 aid_stations -0.0139 -0.00512 0.00607 0.05 percentile
## 3 avg_elevation_gain -0.0554 -0.0515 -0.0465 0.05 percentile
## 4 distance -0.0430 -0.0261 -0.00765 0.05 percentileThe key components are bootstraps() and mutate + map which give you quite a bit of flexibility to compute many statistics.