6.1 Create a Model
6.1.1 Different Model Interfaces

Different-interfaces
- Model Interfaces
- Different Implementations = Different Interfaces
- Linear Regression can be implemented in many ways
- Ordinary Least Squares
- Regularized Linear Regression
- …
- {stats}
- takes formula
- uses
data.frame
lm(formula, data, ...)- {glmnet}
- Has x/y interface
- Uses a matrix
glmnet(x = matrix, y = vector, family = "gaussian", ...)
6.1.2 Model Specification
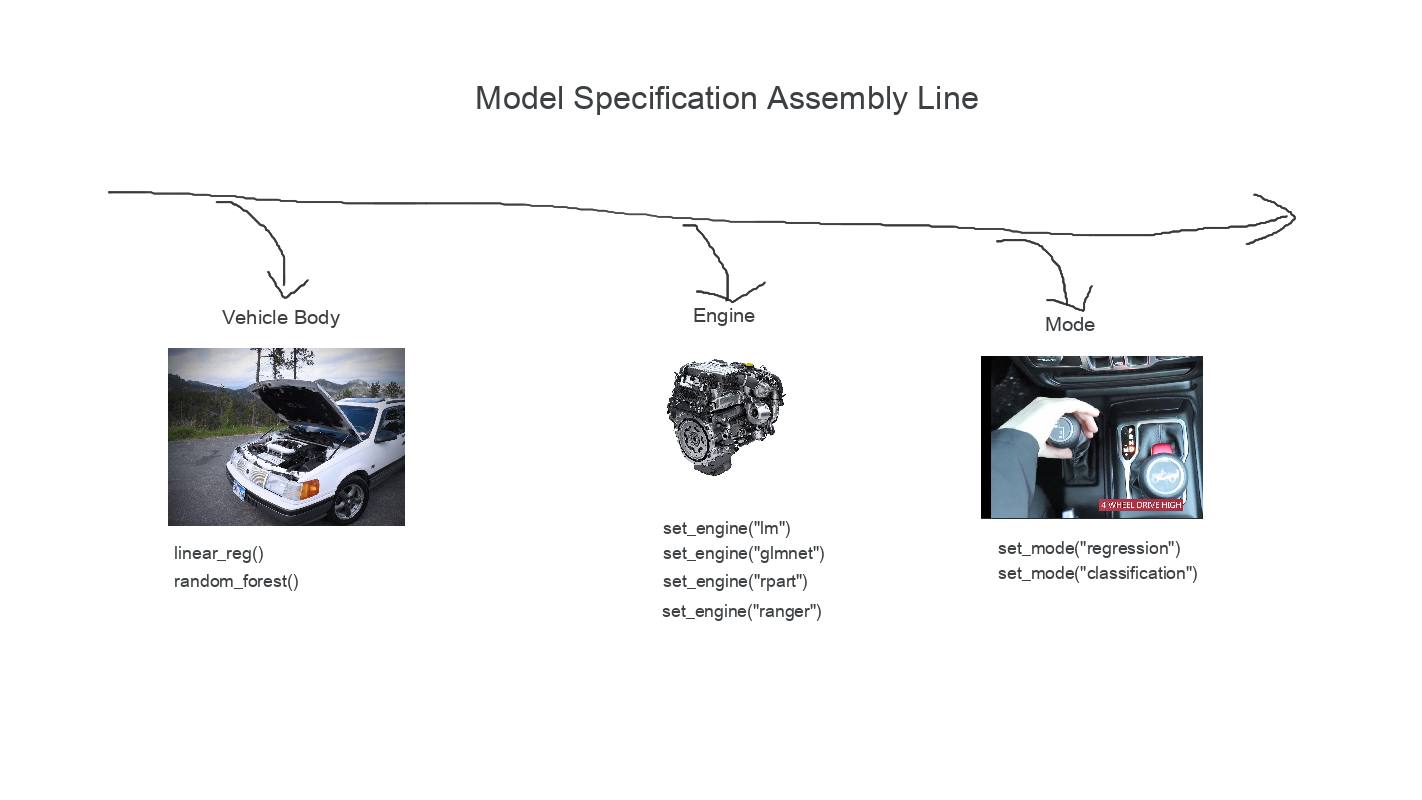
model specification
- {tidymodels}/{parsnip} - Philosophy is to unify & make interfaces more predictable.
- Specify model type (e.g. linear regression, random forest …)
linear_reg()rand_forest()
- Specify engine (i.e. package implementation of algorithm)
set_engine("some package's implementation")
- declare mode (e.g. classification vs linear regression)
- use this when model can do both classification & regression
set_mode("regression")set_mode("classification")
- Specify model type (e.g. linear regression, random forest …)
- Bringing it all together
lm_model_spec <-
linear_reg() %>% # specify model
set_engine("lm") # set engine
lm_model_spec## Linear Regression Model Specification (regression)
##
## Computational engine: lm
6.1.3 Model Fitting
From above we will use our existing model specification
fit()- any nominal or categorical variables will be split out into dummy columns
- most formula methods also turn do the same thing
fit_xy- delays creating dummy variable and has underlying model function
# create model fit using formula
lm_form_fit <-
lm_model_spec %>%
fit(Sale_Price ~ Longitude + Latitude, data = ames_train)
# create model fit using x/y
lm_xy_fit <-
lm_model_spec %>%
fit_xy(
x = ames_train %>% select(Longitude, Latitude),
y = ames_train %>% pull(Sale_Price)
)
6.1.4 Generalized Model Arguments
- Like the varying interfaces, model parameters differ from implementation to implementation
- two level of model arguments
- main arguments - Parameters aligned with the mathematical vehicle
- engine arguments - Parameters aligned with the package implementation of the mathematical algorithm
| argument | ranger | randomForest | sparklyr |
|---|---|---|---|
| sampled predictors | mtry | mtry | feature_subset_strategy |
| trees | num.tress | ntree | num_trees |
| data points to split | min.node.size | nodesize | min_instances_per_node |
| argument | parsnip |
|---|---|
| sampled predictors | mtry |
| trees | trees |
| data points to split | min_n |

Parsnip in Action
- The
translate()provides the mapping from the parsnips interface to the each individual package’s implementation of the algorithm.
# stats implementation
linear_reg() %>%
set_engine("lm") %>%
translate()## Linear Regression Model Specification (regression)
##
## Computational engine: lm
##
## Model fit template:
## stats::lm(formula = missing_arg(), data = missing_arg(), weights = missing_arg())# glmnet implementation
linear_reg(penalty = 1) %>%
set_engine("glmnet") %>%
translate()## Linear Regression Model Specification (regression)
##
## Main Arguments:
## penalty = 1
##
## Computational engine: glmnet
##
## Model fit template:
## glmnet::glmnet(x = missing_arg(), y = missing_arg(), weights = missing_arg(),
## family = "gaussian")