19.1 Equivocal Results
What the heck is this? Loosely, equivacol results are a range of results indicating that the prediction should not be reported.
Let’s take a soccer example. (Data courtesy of 538.)
df <-
here::here('data', 'spi_matches_latest.csv') %>%
read_csv() %>%
drop_na(score1) %>%
mutate(w1 = ifelse(score1 > score2, 'yes', 'no') %>% factor()) %>%
select(
w1, season, matches('(team|spi|prob|importance)[12]'), probtie
) %>%
# we need this as feature, so it can't be NA
drop_na(importance1)# much more data in 2021 season than in older seasons, so use older seasons as test set
trn <- df %>% filter(season == 2021) %>% select(-season)
tst <- df %>% filter(season != 2021) %>% select(-season)
trn %>% count(w1)## # A tibble: 2 × 2
## w1 n
## <fct> <int>
## 1 no 1376
## 2 yes 1054tst %>% count(w1)## # A tibble: 2 × 2
## w1 n
## <fct> <int>
## 1 no 248
## 2 yes 227fit <-
logistic_reg() %>%
set_engine('glm') %>%
# 2 variables cuz dumb humans like 2-D plots
fit(w1 ~ spi1 + importance1, data = trn)
fit %>% tidy()## # A tibble: 3 × 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) -0.742 0.107 -6.92 4.50e-12
## 2 spi1 0.0100 0.00259 3.86 1.13e- 4
## 3 importance1 0.00381 0.00250 1.52 1.27e- 1predict_stuff <- function(fit, set) {
bind_cols(
fit %>%
predict(set),
fit %>%
predict(set, type = 'prob'),
fit %>%
predict(set, type = 'conf_int', std_error = TRUE),
set
)
}
preds_tst <- fit %>% predict_stuff(tst)How’s the model accuracy?
preds_tst %>%
accuracy(estimate = .pred_class, truth = w1)## # A tibble: 1 × 3
## .metric .estimator .estimate
## <chr> <chr> <dbl>
## 1 accuracy binary 0.579Seems reasonable… so maybe modeling soccer match outcomes with 2 variables ain’t so bad, eh?
Observe the fitted class boundary. (Confidence intervals shown instead of prediction intervals because I didn’t want to use stan… Max please forgive me.)
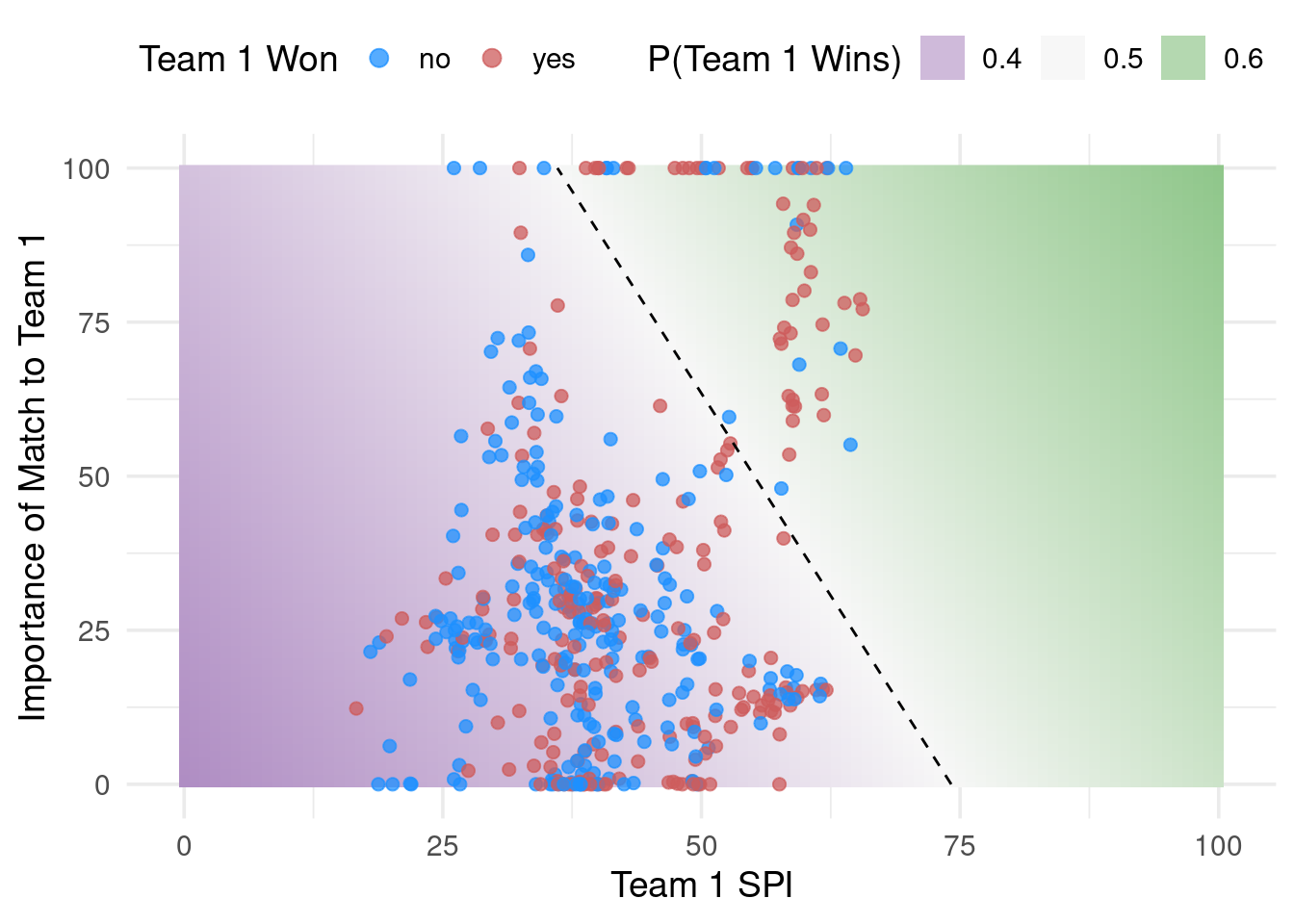
Use the {probably} package!
lvls <- levels(preds_tst$w1)
preds_tst_eqz <-
preds_tst %>%
mutate(.pred_with_eqz = make_two_class_pred(.pred_yes, lvls, buffer = 0.025))
preds_tst_eqz %>% count(.pred_with_eqz)## # A tibble: 3 × 2
## .pred_with_eqz n
## <clss_prd> <int>
## 1 [EQ] 50
## 2 no 48
## 3 yes 377Look at how make_two_class_pred changes our confusion matrix.
# All data
preds_tst_eqz %>% conf_mat(w1, .pred_class) %>% autoplot('heatmap')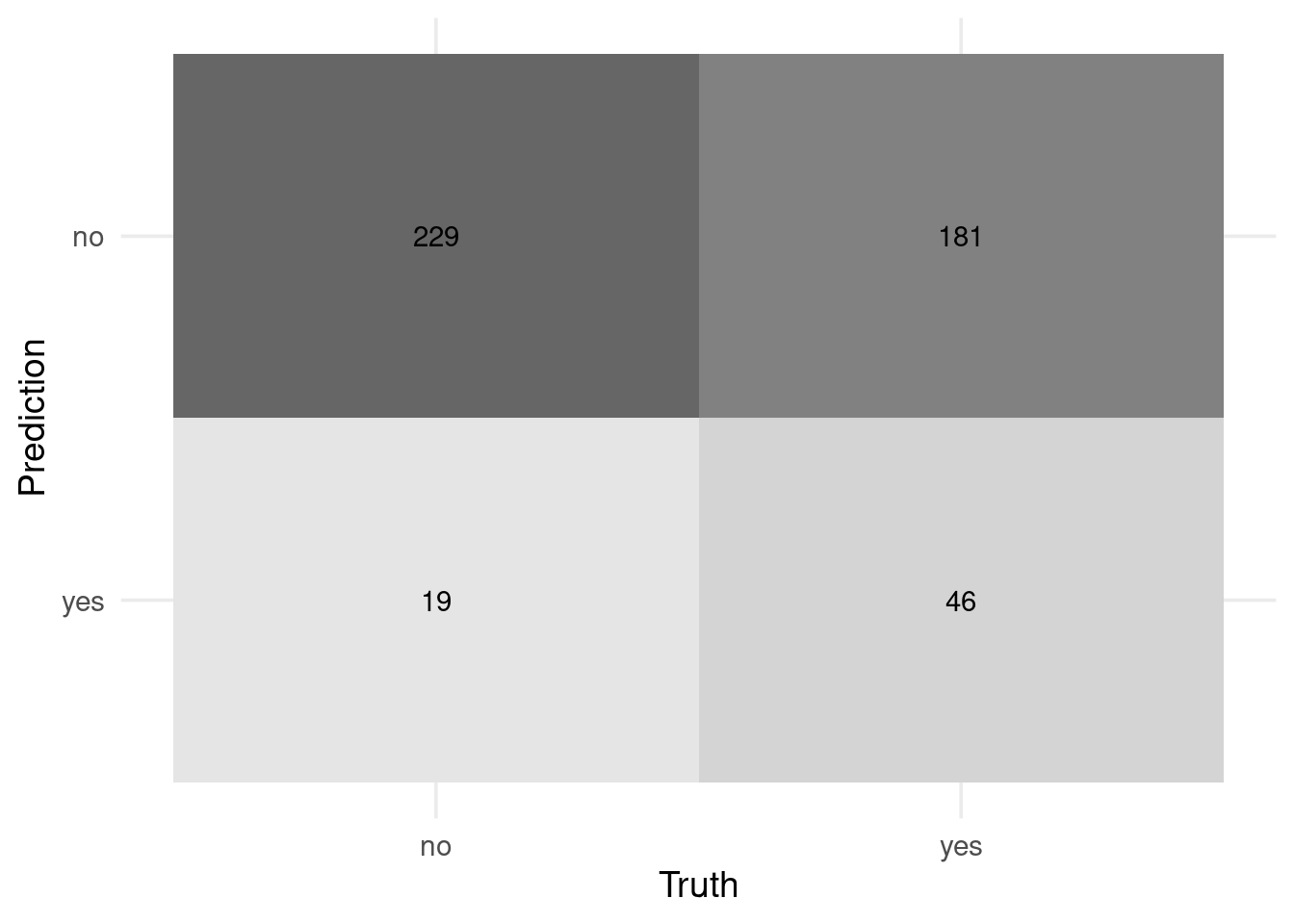
# Reportable results only
preds_tst_eqz %>% conf_mat(w1, .pred_with_eqz) %>% autoplot('heatmap')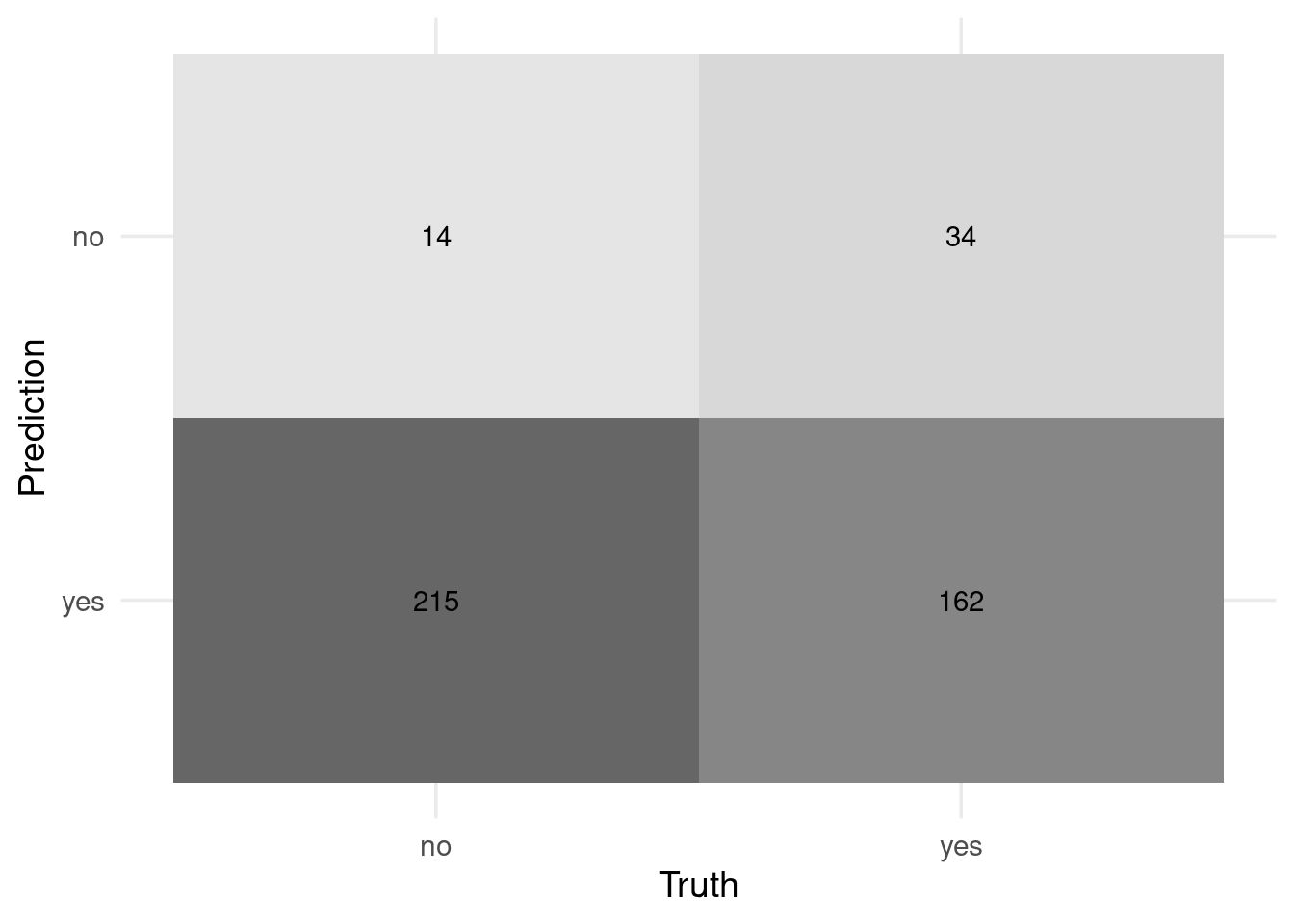
Does the equivocal zone help improve accuracy? How sensitive is accuracy and our reportable rate to the width of the buffer?
eq_zone_results <- function(buffer) {
preds_tst_eqz <-
preds_tst %>%
mutate(.pred_with_eqz = make_two_class_pred(.pred_no, lvls, buffer = buffer))
acc <- preds_tst_eqz %>% accuracy(w1, .pred_with_eqz)
rep_rate <- reportable_rate(preds_tst_eqz$.pred_with_eqz)
tibble(accuracy = acc$.estimate, reportable = rep_rate, buffer = buffer)
}
map_dfr(seq(0, 0.15, length.out = 40), eq_zone_results) %>%
pivot_longer(c(-buffer)) %>%
ggplot(aes(x = buffer, y = value, col = name)) +
geom_step(size = 1.2, alpha = 0.8) +
labs(y = NULL)
How does the standard error look across the feature space

Makes sense… we’re more uncertain for cases outside of the normal boundary of our data.