- Setup parallel processing to speed up the tuning execution time
library(doParallel)## Loading required package: foreach##
## Attaching package: 'foreach'## The following objects are masked from 'package:purrr':
##
## accumulate, when## Loading required package: iterators## Loading required package: parallelall_cores <- parallel::detectCores(logical = FALSE)
registerDoParallel(cores = all_cores - 1)- Execute the hyperparameter tuning
set.seed(345)
tree_rs <- tune_grid(
wflw_tree,
resamples = folds,
grid = tree_grid,
control = control_grid(
save_pred = TRUE
)
)
tree_rs## # Tuning results
## # 3-fold cross-validation using stratification
## # A tibble: 3 × 5
## splits id .metrics .notes .predictions
## <list> <chr> <list> <list> <list>
## 1 <split [734/367]> Fold1 <tibble [250 × 7]> <tibble [0 × 3]> <tibble>
## 2 <split [734/367]> Fold2 <tibble [250 × 7]> <tibble [0 × 3]> <tibble>
## 3 <split [734/367]> Fold3 <tibble [250 × 7]> <tibble [0 × 3]> <tibble>- Evaluate the model
# plot evaluation metrics
tree_rs %>%
autoplot()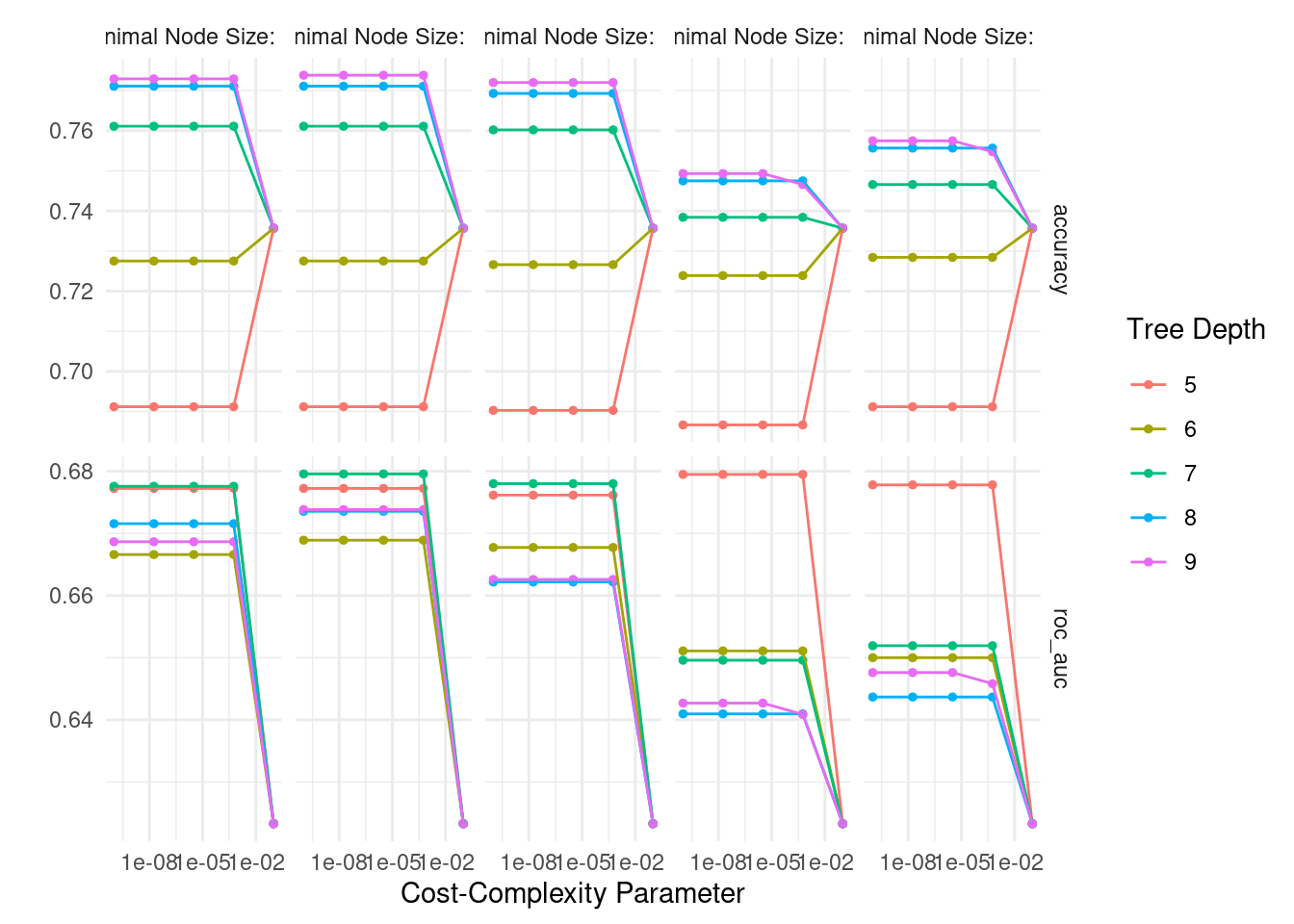
# sort `roc_auc` metric (descending)
tree_rs %>%
collect_metrics() %>%
filter(.metric == "roc_auc") %>%
arrange(-mean)## # A tibble: 125 × 9
## cost_complexity tree_depth min_n .metric .estimator mean n std_err
## <dbl> <int> <int> <chr> <chr> <dbl> <int> <dbl>
## 1 0.0000000001 7 12 roc_auc binary 0.680 3 0.0388
## 2 0.0000000178 7 12 roc_auc binary 0.680 3 0.0388
## 3 0.00000316 7 12 roc_auc binary 0.680 3 0.0388
## 4 0.000562 7 12 roc_auc binary 0.680 3 0.0388
## 5 0.0000000001 5 14 roc_auc binary 0.679 3 0.0260
## 6 0.0000000178 5 14 roc_auc binary 0.679 3 0.0260
## 7 0.00000316 5 14 roc_auc binary 0.679 3 0.0260
## 8 0.000562 5 14 roc_auc binary 0.679 3 0.0260
## 9 0.0000000001 7 13 roc_auc binary 0.678 3 0.0403
## 10 0.0000000178 7 13 roc_auc binary 0.678 3 0.0403
## # ℹ 115 more rows
## # ℹ 1 more variable: .config <chr>Show best model
tree_rs %>%
show_best(metric = 'roc_auc')## # A tibble: 5 × 9
## cost_complexity tree_depth min_n .metric .estimator mean n std_err
## <dbl> <int> <int> <chr> <chr> <dbl> <int> <dbl>
## 1 0.0000000001 7 12 roc_auc binary 0.680 3 0.0388
## 2 0.0000000178 7 12 roc_auc binary 0.680 3 0.0388
## 3 0.00000316 7 12 roc_auc binary 0.680 3 0.0388
## 4 0.000562 7 12 roc_auc binary 0.680 3 0.0388
## 5 0.0000000001 5 14 roc_auc binary 0.679 3 0.0260
## # ℹ 1 more variable: .config <chr>- Choose the best model hyperparameters and create the last workflow and last fit(fit with the full train dataset and evaluate with the test dataset)
# select best model by 'roc_auc'
best_tree_roc_auc <- select_best(tree_rs, "roc_auc")
best_tree_roc_auc## # A tibble: 1 × 4
## cost_complexity tree_depth min_n .config
## <dbl> <int> <int> <chr>
## 1 0.0000000001 7 12 Preprocessor1_Model036# create final workflow
final_tree <- finalize_workflow(
wflw_tree,
best_tree_roc_auc
)
# last fit
final_fit_tree <- last_fit(
final_tree,
churn_split
)
final_fit_tree %>%
collect_metrics()## # A tibble: 2 × 4
## .metric .estimator .estimate .config
## <chr> <chr> <dbl> <chr>
## 1 accuracy binary 0.726 Preprocessor1_Model1
## 2 roc_auc binary 0.616 Preprocessor1_Model1- Feature importance
# first 20 most important features
final_fit_tree %>%
extract_fit_parsnip() %>%
vip(num_features = 20, geom = 'point')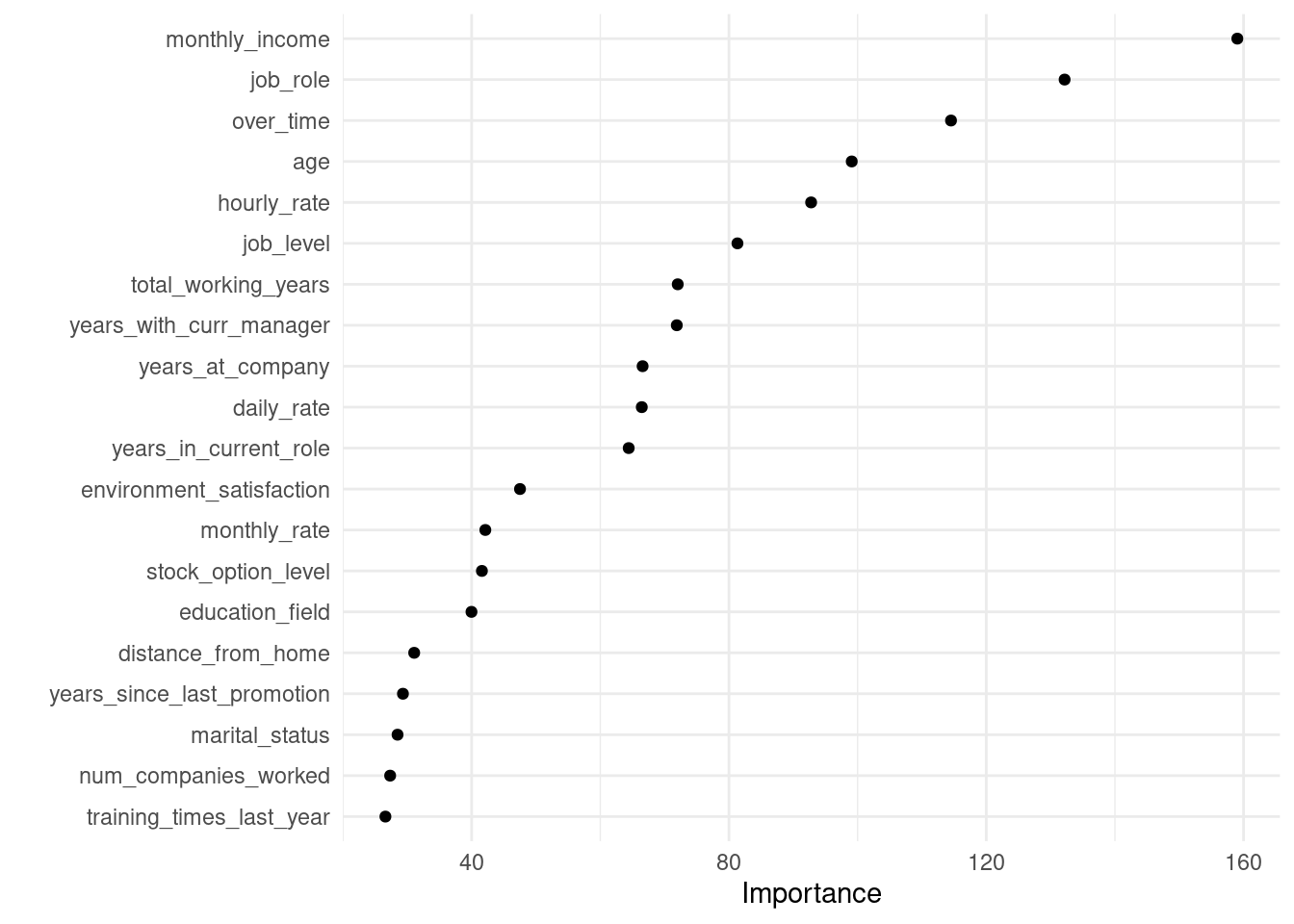
- Generate predictions
# predictions and confusion matrix
final_fit_tree %>%
collect_predictions() %>%
conf_mat(attrition, .pred_class) %>%
pluck(1) %>%
as_tibble() %>%
ggplot(aes(Prediction, Truth, alpha = n)) +
geom_tile(show.legend = FALSE) +
geom_text(aes(label = n), colour = "white", alpha = 1, size = 8)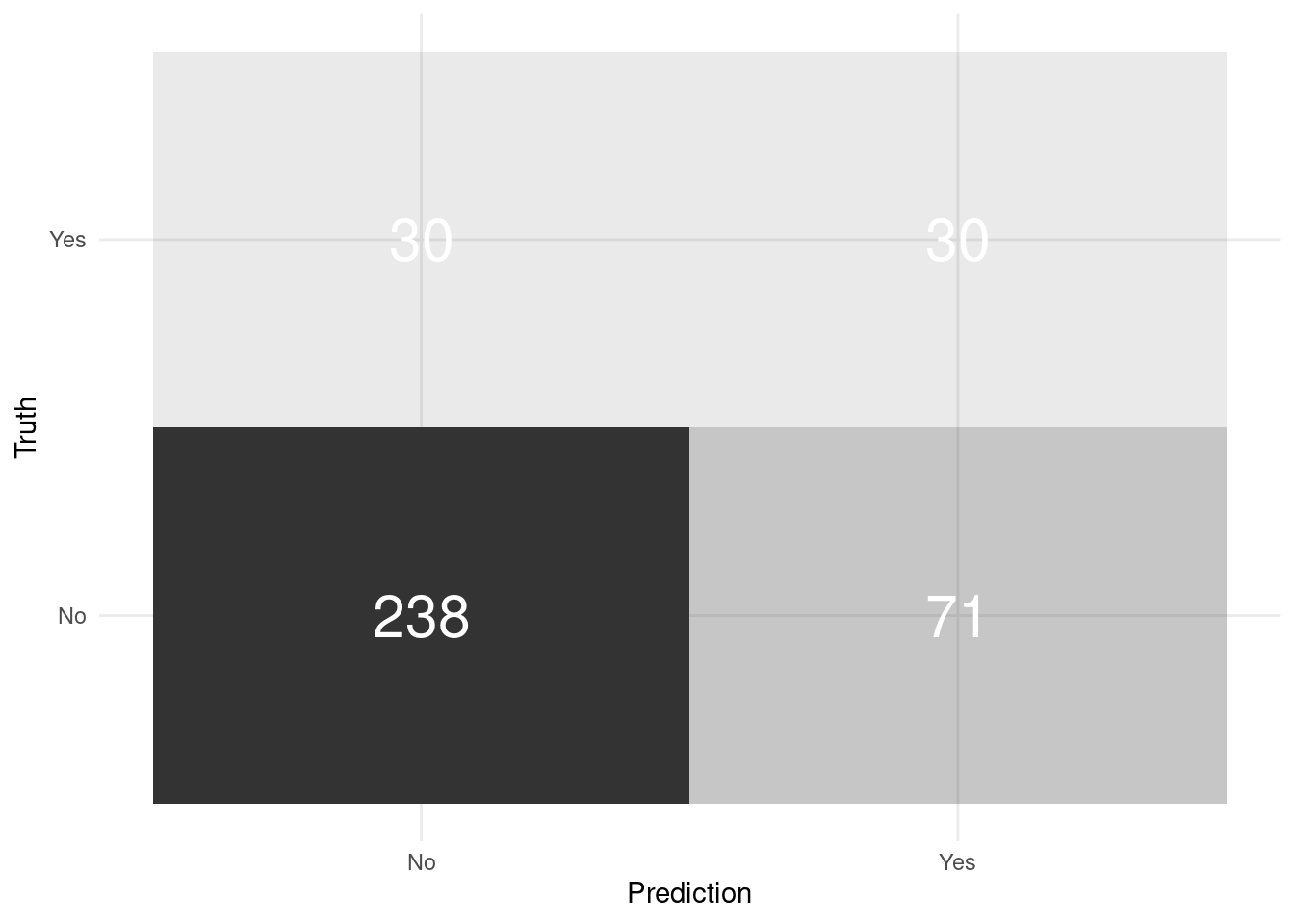
# F1 metrics (final model)
final_fit_tree %>%
collect_predictions() %>%
f_meas(attrition, .pred_class) %>%
select(-.estimator)## # A tibble: 1 × 2
## .metric .estimate
## <chr> <dbl>
## 1 f_meas 0.825# ROC curve
final_fit_tree %>%
collect_predictions() %>%
roc_curve(attrition, .pred_No) %>%
autoplot()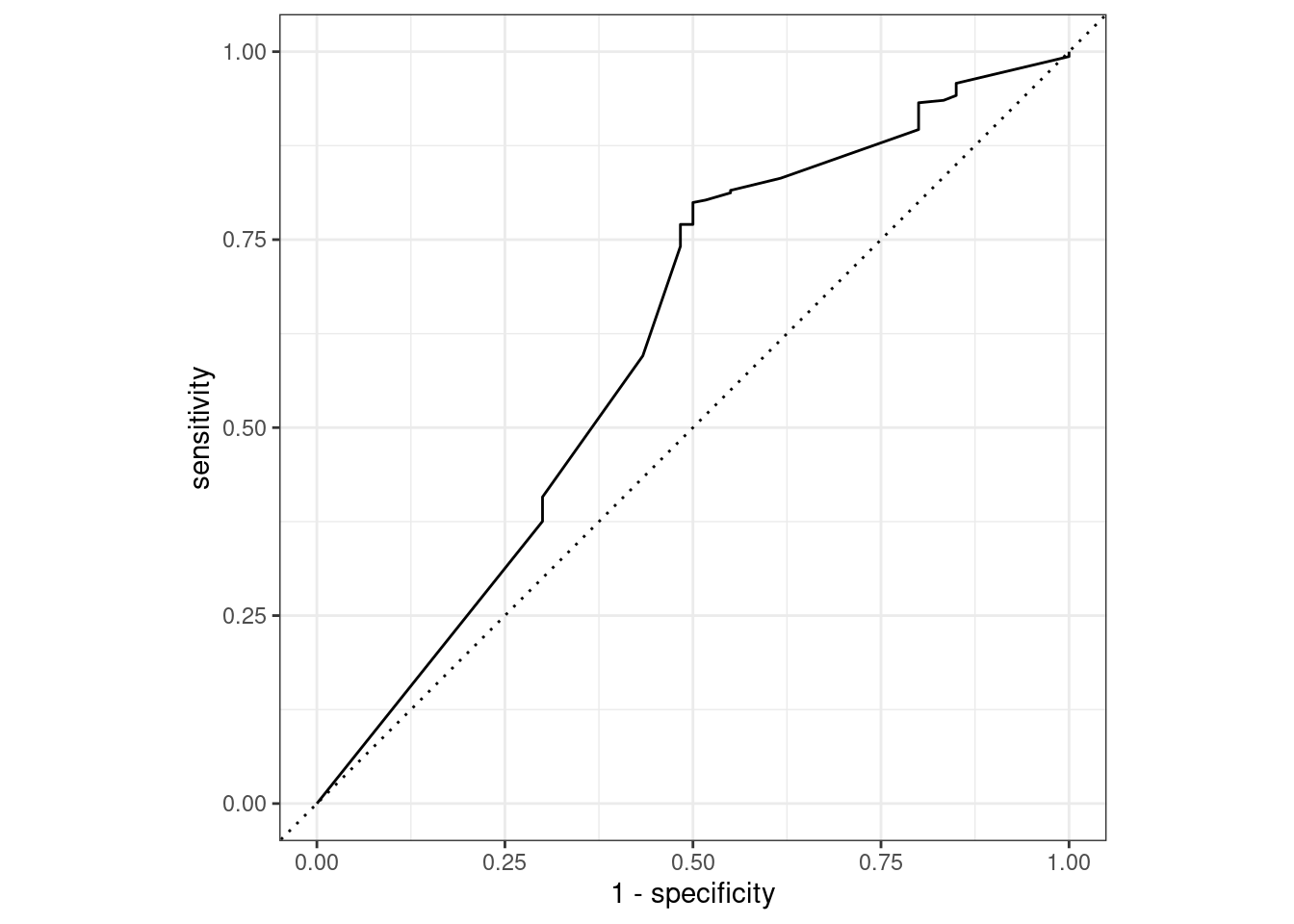
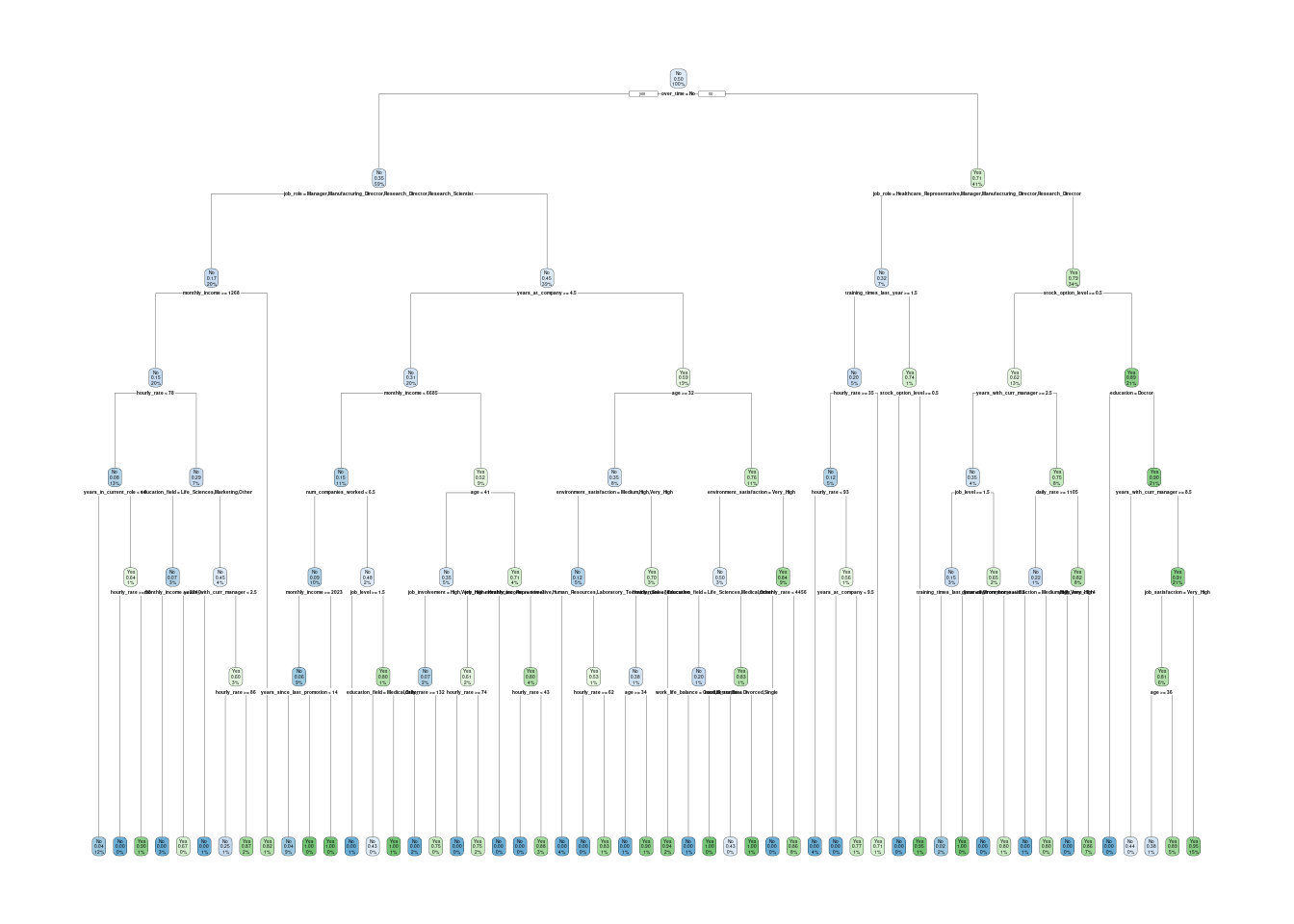
# plot `rpart` final tree
tree_fit_rpart <- final_fit_tree %>%
extract_fit_engine(final_tree)
rpart.plot(tree_fit_rpart)## Warning: Cannot retrieve the data used to build the model (so cannot determine roundint and is.binary for the variables).
## To silence this warning:
## Call rpart.plot with roundint=FALSE,
## or rebuild the rpart model with model=TRUE.## Warning: labs do not fit even at cex 0.15, there may be some overplotting