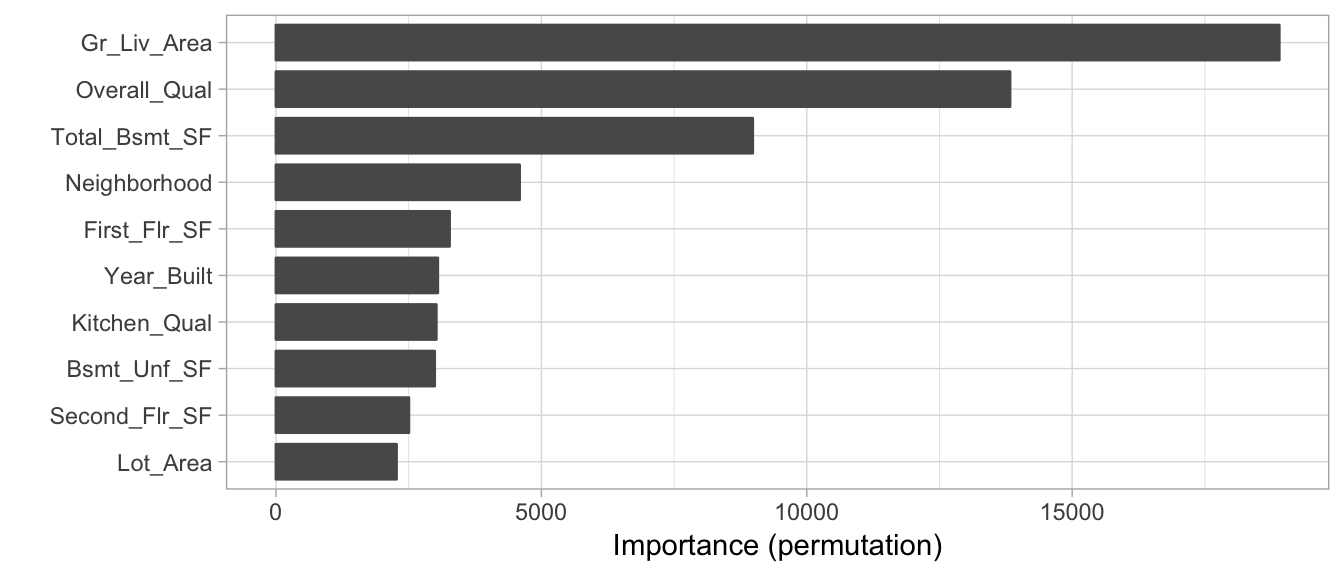
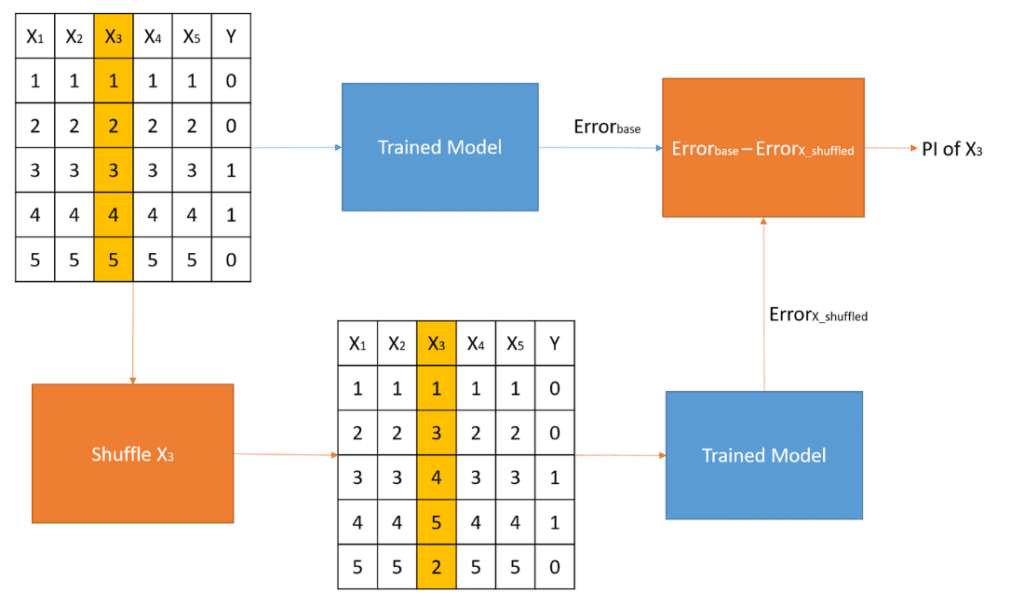
16.5 Permutation-based feature importance
16.5.1 Concept
We can use this method for any model after assuring that:
- The model isn’t overfitting: To avoid interpreting noise rather than signal.
- Features present low correlation: To avoid under estimating the importance of correlated features, as permuting a feature without permuting its correlated pairs won’t have a big effect.
It measures feature’s importance by calculating the increase of the model’s prediction error, by taking the difference to interpret absolute values or the ratio to interpret relative values of a metric like the RMSE after permuting the feature, as describes the next algorithm:
For any given loss function do the following:
1. Compute loss function for original model
2. For variable i in {1,...,p} do
| randomize values
| apply given ML model
| estimate loss function
| compute feature importance (some difference/ratio measure
between permuted loss & original loss)
End
3. Sort variables by descending feature importance16.5.2 Implementation
General Description
| Package | Function | Description |
|---|---|---|
| iml | FeatureImp() |
- Several loss functions (loss = "mse") - Difference and ratio calculations. |
| DALEX | variable_importance() |
- Several loss functions. - Difference and ratio calculations. - It can sample the data ( n_sample = 1000) |
| vip | vip() or vi_permute() |
- Supports model-specific and model-agnostic approaches. - Custom and several loss functions. - Perform a Monte Carlo simulation to stabilize the procedure. ( nsim = 5) - It can sample the data ( sample_size or sample_frac) - It can perform parallel computations ( parallel = TRUE) |
Creating Needed Objects
# For iml and DALEX
# 1) Split the features and response
features <- as.data.frame(train_h2o) |> select(-Sale_Price)
response <- as.data.frame(train_h2o) |> pull(Sale_Price)
# 2) Create custom predict function that returns the predicted values as a vector
pred <- function(object, newdata) {
results <- as.vector(h2o.predict(object, as.h2o(newdata)))
return(results)
}
## For DALEX
components_dalex <- DALEX::explain(
model = ensemble_tree,
data = features,
y = response,
predict_function = pred
) iml
Predictor$new(ensemble_tree, data = features, y = response) |>
FeatureImp$new(loss = "rmse", compare = "difference") |>
plot()DALEX
loss_default(components_dalex$model_info$type)
model_parts(components_dalex,
type = "difference") |>
plot()vip
vip(
ensemble_tree,
train = as.data.frame(train_h2o),
method = "permute",
target = "Sale_Price",
metric = "RMSE",
type = "difference",
nsim = 5,
sample_frac = 0.5,
pred_wrapper = pred
)