Aside on log transformations
Note: This following material was not discussed in the EMA book.
Fitting a regression model on a log transformed target variable, then applying the inverse operation on the logged model prediction will result in biased predictions. This may or may not be a problem depending on your goals, loss functions, etc.
Helpful links:
Refer to the references below regarding techniques for bias correction when logging the target variable:
- Duan’s transformation: https://stats.stackexchange.com/questions/55692/back-transformation-of-an-mlr-model
- Example of Duan’s smearing in python: https://andrewpwheeler.com/tag/linear-regression/
- Alternative smearing adjustment using regression without an intercept: https://stats.stackexchange.com/questions/361618/how-to-back-transform-a-log-transformed-regression-model-in-r-with-bias-correcti
- Good discussion on target variable transformation: https://florianwilhelm.info/2020/05/honey_i_shrunk_the_target_variable/
- Lognormal smearing: https://en.wikipedia.org/wiki/Smearing_retransformation
- Forecasting bias: https://arxiv.org/pdf/2208.12264
Example Bias Adjustment factors using Duan’s Transformation
Duan’s transformation Formula using log base 10:
\[\hat{Y}_j = 10^{(\widehat{\log_{10}{Y}}_j)} \cdot \frac{1}{N}\sum_{i=1}^N 10^{e_i}\]
where \(\widehat{\log_{10}{Y}}_j\) is the model’s prediction on the logged scale, and \(e_i\) is the model residual on the logged scale (i.e. \(log_{10} Y - \widehat{\log_{10}{Y}}_j\)).
ols smearing adjustment: 1.032rf smearing adjustment: 1.008gbm shallow smearing adjustment: 1.049gbm deep smearing adjustment: 1.02Below is a comparison of model performance on our test set using three different modeling methodologies:
- Using the raw scale target variable for modeling
- Using the log transformed target for modeling, then taking the inverse
- Using the log transformed target for modeling, then taking the inverse * smearing adjustment
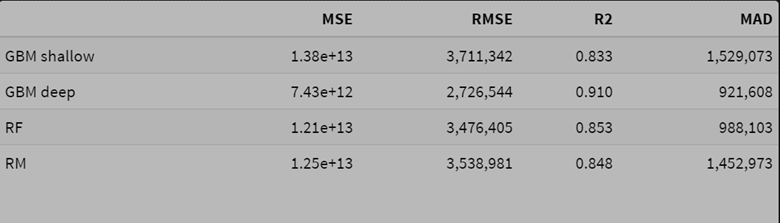
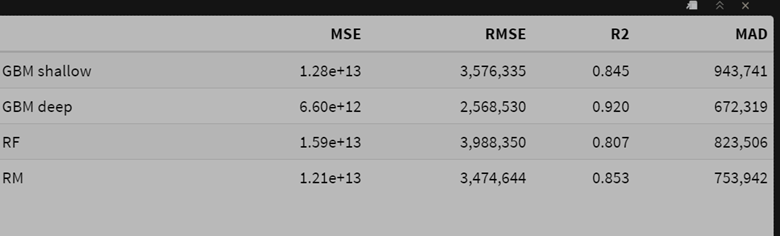
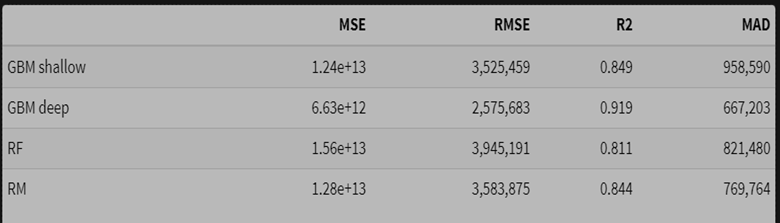
Observations:
- The random forest model performs best in terms of squared error on the raw scale.
- The smearing adjustment improves the random forest compared to the basic back-transformed log model.
- The deep GBM model’s MAD value improves slightly with smearing, with a very small deterioration in squared error.