3.4 Attributes
Attributes are name-value pairs that attach metadata to an object(vector).
- Name-value pairs: attributes have a name and a value
- Metadata: not data itself, but data about the data
3.4.1 How?
3.4.1.1 Getting and Setting
Three functions:
- retrieve and modify single attributes with
attr() - retrieve en masse with
attributes() - set en masse with
structure()
Single attribute
Use attr()
# some object
a <- c(1, 2, 3)
# set attribute
attr(x = a, which = "attribute_name") <- "some attribute"
# get attribute
attr(a, "attribute_name")
#> [1] "some attribute"Multiple attributes
To set multiple attributes, use structure() To get multiple attributes, use attributes()
a <- 1:3
attr(a, "x") <- "abcdef"
attr(a, "x")
#> [1] "abcdef"
attr(a, "y") <- 4:6
str(attributes(a))
#> List of 2
#> $ x: chr "abcdef"
#> $ y: int [1:3] 4 5 6
# Or equivalently
a <- structure(
1:3,
x = "abcdef",
y = 4:6
)
str(attributes(a))
#> List of 2
#> $ x: chr "abcdef"
#> $ y: int [1:3] 4 5 6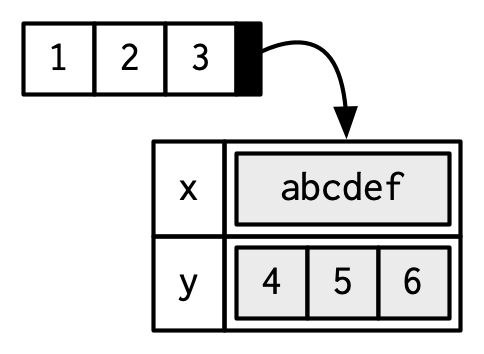
3.4.2 Why
Three particularly important attributes:
- names - a character vector giving each element a name
- dimension - (or dim) turns vectors into matrices and arrays
- class - powers the S3 object system (we’ll learn more about this in chapter 13)
Most attributes are lost by most operations. Only two attributes are routinely preserved: names and dimension.
3.4.2.1 Names
Three Four ways to name:
# (1) When creating it:
x <- c(A = 1, B = 2, C = 3)
x
#> A B C
#> 1 2 3
# (2) By assigning a character vector to names()
y <- 1:3
names(y) <- c("a", "b", "c")
y
#> a b c
#> 1 2 3
# (3) Inline, with setNames():
z <- setNames(1:3, c("a", "b", "c"))
z
#> a b c
#> 1 2 3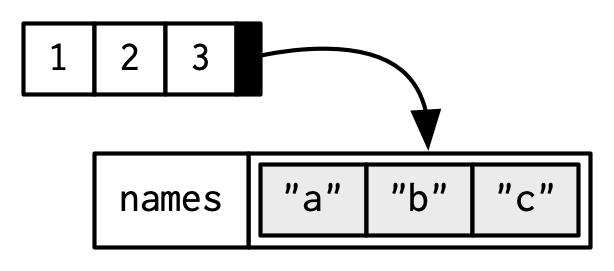
# (4) By setting names--with {rlang}
a <- 1:3
rlang::set_names(
x = a,
nm = c("a", "b", "c")
)
#> a b c
#> 1 2 3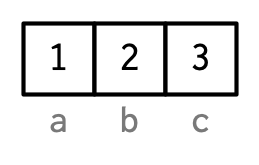
- You can remove names from a vector by using
x <- unname(x)ornames(x) <- NULL. - Thematically but not directly related: labelled class vectors with
haven::labelled()
3.4.2.2 Dimensions
Create matrices and arrays with matrix() and array(), or by using the assignment form of dim():
# Two scalar arguments specify row and column sizes
x <- matrix(1:6, nrow = 2, ncol = 3)
x
#> [,1] [,2] [,3]
#> [1,] 1 3 5
#> [2,] 2 4 6
# One vector argument to describe all dimensions
y <- array(1:24, c(2, 3, 4)) # rows, columns, no of arrays
y
#> , , 1
#>
#> [,1] [,2] [,3]
#> [1,] 1 3 5
#> [2,] 2 4 6
#>
#> , , 2
#>
#> [,1] [,2] [,3]
#> [1,] 7 9 11
#> [2,] 8 10 12
#>
#> , , 3
#>
#> [,1] [,2] [,3]
#> [1,] 13 15 17
#> [2,] 14 16 18
#>
#> , , 4
#>
#> [,1] [,2] [,3]
#> [1,] 19 21 23
#> [2,] 20 22 24
# You can also modify an object in place by setting dim()
z <- 1:6
dim(z) <- c(2, 3) # rows, columns
z
#> [,1] [,2] [,3]
#> [1,] 1 3 5
#> [2,] 2 4 6
a <- 1:24
dim(a) <- c(2, 3, 4) # rows, columns, no of arrays
a
#> , , 1
#>
#> [,1] [,2] [,3]
#> [1,] 1 3 5
#> [2,] 2 4 6
#>
#> , , 2
#>
#> [,1] [,2] [,3]
#> [1,] 7 9 11
#> [2,] 8 10 12
#>
#> , , 3
#>
#> [,1] [,2] [,3]
#> [1,] 13 15 17
#> [2,] 14 16 18
#>
#> , , 4
#>
#> [,1] [,2] [,3]
#> [1,] 19 21 23
#> [2,] 20 22 243.4.2.2.1 Functions for working with vectors, matrices and arrays:
| Vector | Matrix | Array |
|---|---|---|
names() |
rownames(), colnames() |
dimnames() |
length() |
nrow(), ncol() |
dim() |
c() |
rbind(), cbind() |
abind::abind() |
| — | t() |
aperm() |
is.null(dim(x)) |
is.matrix() |
is.array() |
- Caution: A vector without a
dimattribute set is often thought of as 1-dimensional, but actually hasNULLdimensions. - One dimension?
3.4.3 Exercises
- How is
setNames()implemented? How isunname()implemented? Read the source code.
Answer(s)
setNames() is implemented as:
Because the data argument comes first, setNames() also works well with the magrittr-pipe operator. When no first argument is given, the result is a named vector (this is rather untypical as required arguments usually come first):
unname() is implemented in the following way:
unname <- function(obj, force = FALSE) {
if (!is.null(names(obj)))
names(obj) <- NULL
if (!is.null(dimnames(obj)) && (force || !is.data.frame(obj)))
dimnames(obj) <- NULL
obj
}unname() removes existing names (or dimnames) by setting them to NULL.
- What does
dim()return when applied to a 1-dimensional vector? When might you useNROW()orNCOL()?
Answer(s)
dim() will return NULL when applied to a 1d vector.
One may want to use NROW() or NCOL() to handle atomic vectors, lists and NULL values in the same way as one column matrices or data frames. For these objects nrow() and ncol() return NULL:
- How would you describe the following three objects? What makes them different from
1:5?
Answer(s)
- An early draft used this code to illustrate
structure():
But when you print that object you don’t see the comment attribute. Why? Is the attribute missing, or is there something else special about it?
Answer(s)
The documentation states (see ?comment):
Contrary to other attributes, the comment is not printed (by print or print.default).
Also, from ?attributes:
Note that some attributes (namely class, comment, dim, dimnames, names, row.names and tsp) are treated specially and have restrictions on the values which can be set.
We can retrieve comment attributes by calling them explicitly: