10.4 Example: Visualising Lung Cancer Deaths by Prevalence and Age in Germany
## # A tibble: 6 × 8
## age sex prevalence prev_upper prev_lower dx dx_uppe dx_lower
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 10-14 male 0.08 0.13 0.05 0.322 0.461 0.217
## 2 10-14 female 0.18 0.32 0.09 0.457 0.761 0.248
## 3 10-14 both 0.13 0.22 0.07 0.779 1.21 0.468
## 4 15-19 male 0.48 0.77 0.29 1.27 1.75 0.916
## 5 15-19 female 0.9 1.52 0.5 1.56 2.46 0.941
## 6 15-19 both 0.68 1.02 0.44 2.83 3.88 2.07Scatter plot
library(ggplot2)
lung_scatter <- hmsidwR::germany_lungc |>
# Initialise ggplot with arguments to set the x and y
# aesthetics
ggplot(aes(x = prevalence, y = dx)) +
# Create a scatter plot, using sex to modify the shape
# aesthetic
geom_point(aes(shape = sex)) +
# Set the title and names for x and y axes
labs(title = "Lung Cancer Deaths by Prevalence in Germany",
x = "Prevalence",
y = "Deaths")
lung_scatter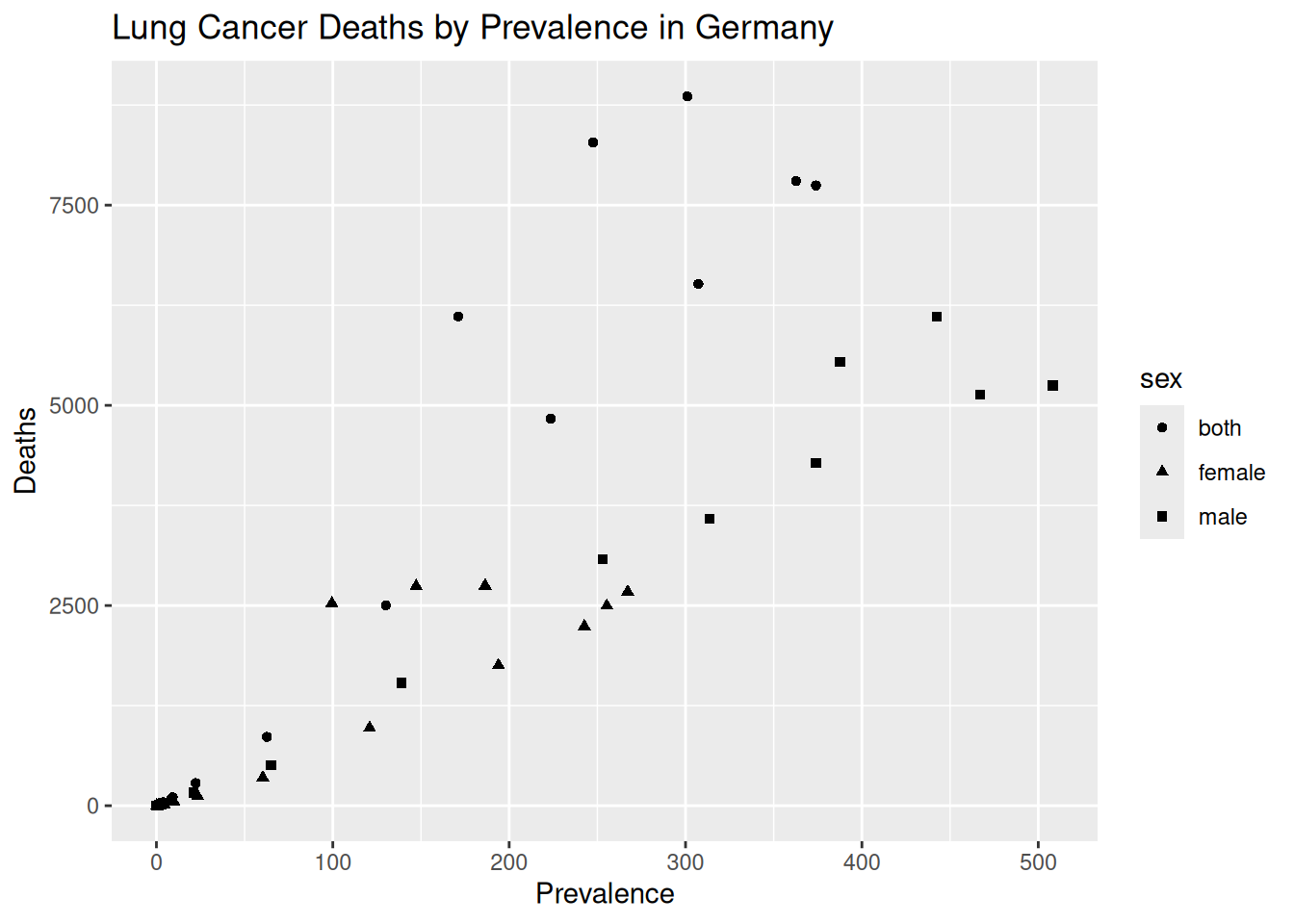
Things to do:
Change the size of the points with the
sizeaesthetic withingeom_point.Change the colour of the points with the
colourargument withingeom_point.Remove the grey background with
theme:theme_classic()ortheme_bw()with extra parameters do the same job.
Check the readability of colours for people with colour vision deficiency.
hmsidwR::germany_lungc |>
ggplot(
aes(
x = prevalence,
y = dx,
shape = sex,
colour = sex)) +
geom_point(size = 4) +
labs(title = "Lung Cancer Deaths by Prevalence in Germany",
x = "Prevalence",
y = "Deaths") +
theme_classic() +
theme(text = element_text(family = "Lato",
size = 14),
axis.text = element_text(colour = "black"))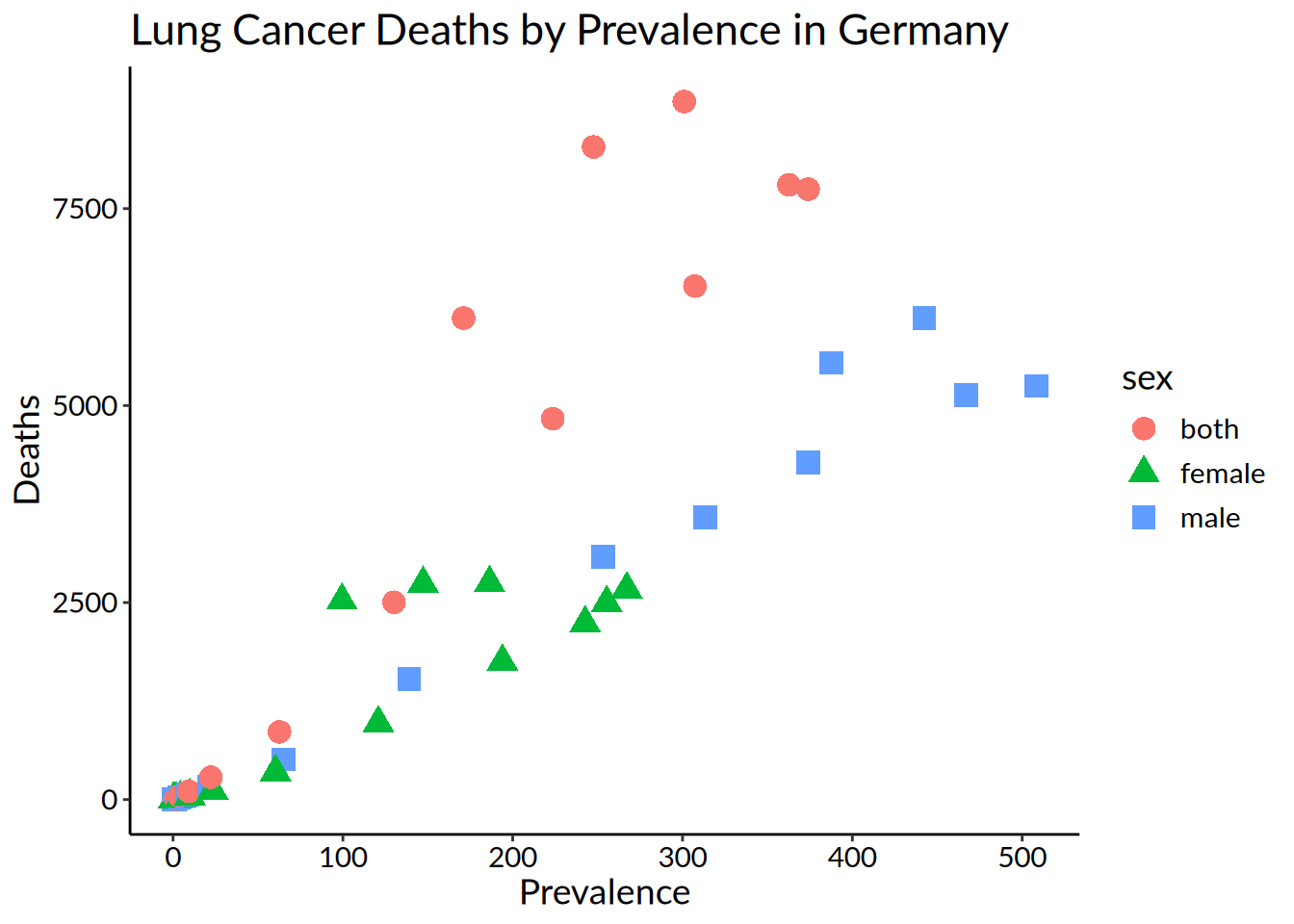
hmsidwR::germany_lungc |>
ggplot(
aes(
x = prevalence,
y = dx,
shape = sex,
colour = sex)) +
geom_point(size = 4) +
labs(title = "Lung Cancer Deaths by Prevalence in Germany",
x = "Prevalence",
y = "Deaths") +
theme_bw() +
theme(axis.line = element_line(colour = "black"),
plot.background = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
text = element_text(family = "Lato",
size = 14),
axis.text = element_text(colour = "black"))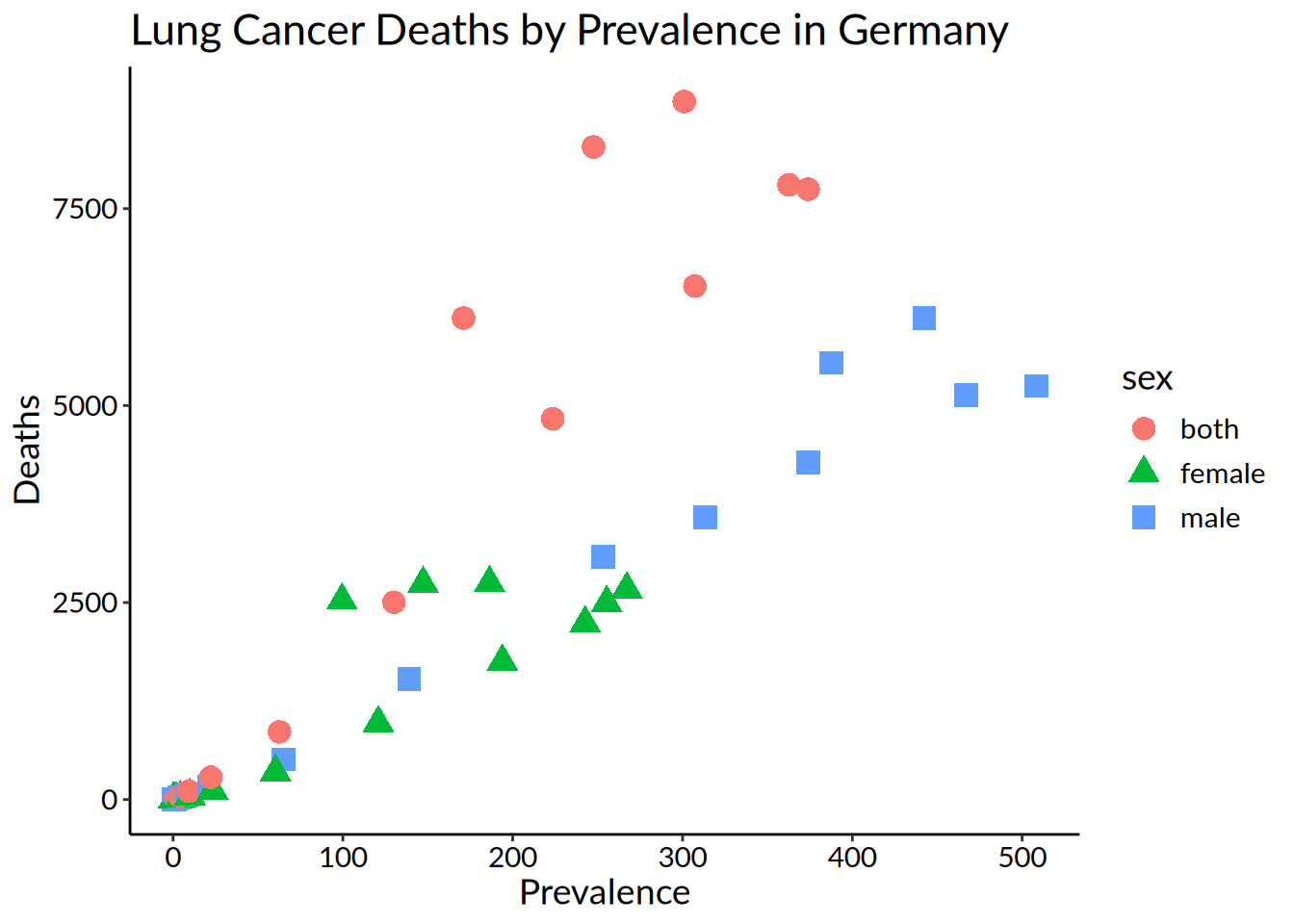
It’s ok, but maybe not for people who are colourblind.
How to check?
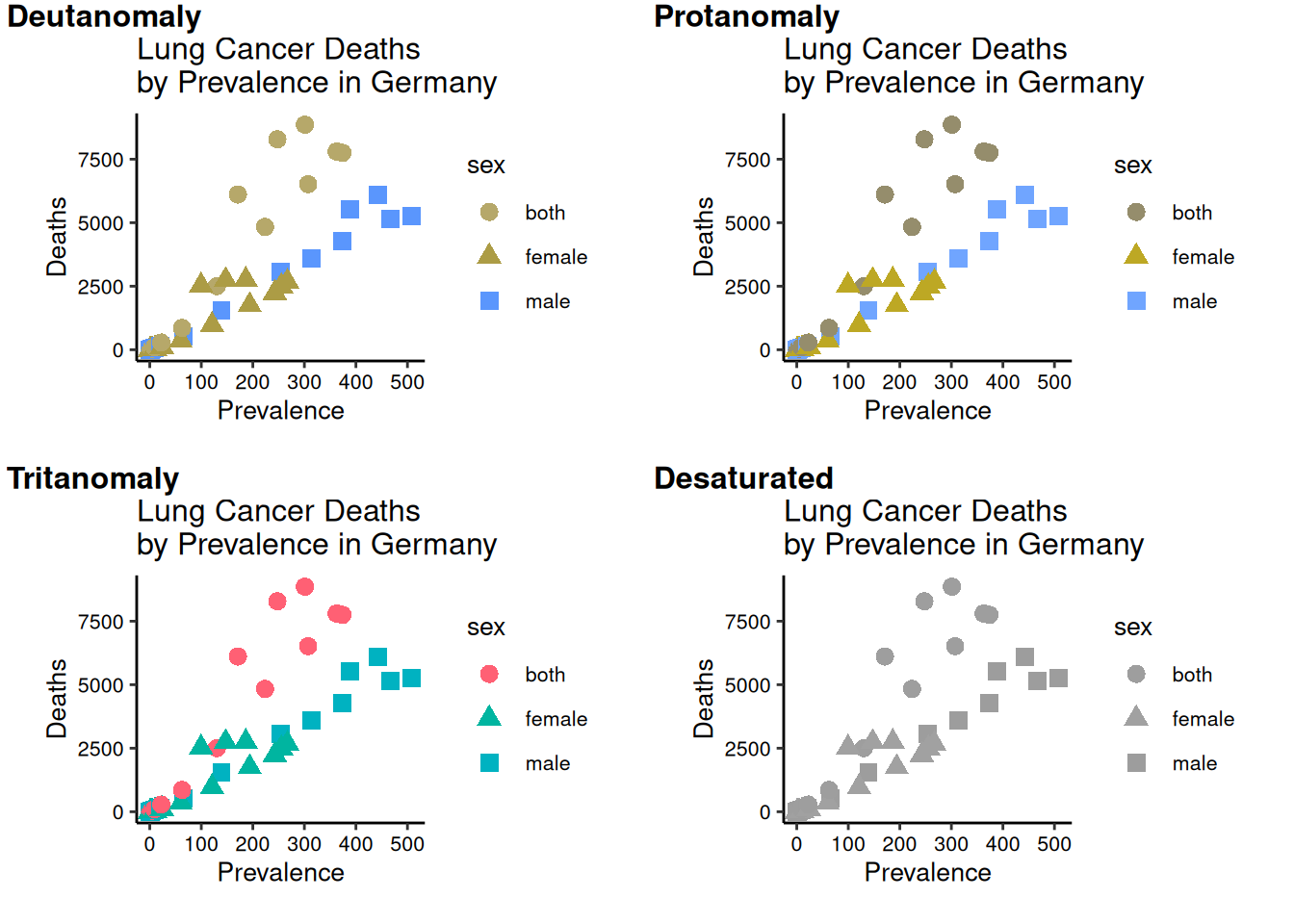
Possible options:
Don’t use colour.
Put black outlines around the shapes.
Choose a better palette.
The Okabe-Ito palette (below) was designed for qualitative data.
The
viridispalette may be suitable for continuous data.{colorbrewer}package has colourblind-friendly palettes.
- Control colour and shape using
scale_colour_manualandscale_shape_manual(necessary if modifying the legend title).
scatter +
labs(title = "Lung Cancer Deaths by Prevalence in Germany") +
scale_color_manual(name = "Sex",
values = c("#000000", "#009E73", "#D55E00"),
labels = c("Both", "Female", "Male")) +
scale_shape_manual(name = "Sex",
values = c(16, 17, 15),
labels = c("Both", "Female", "Male"))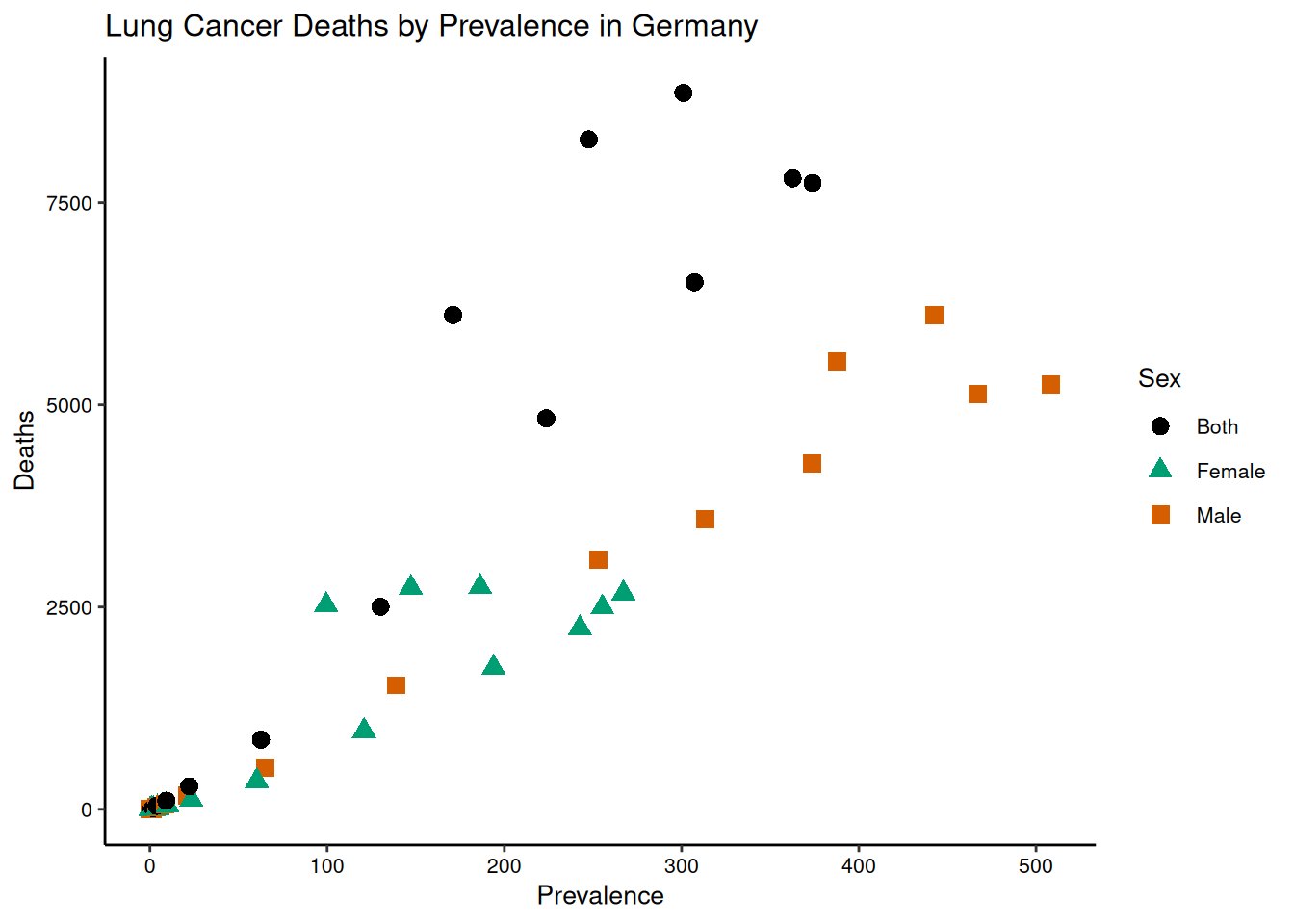
- Can also use
scale_color_OkabeItoandscale_fill_OkabeIto.
Barplot
library(ggpattern)
# Create a barplot
hmsidwR::germany_lungc |>
# This time, the aesthetics are age and estimated death rate
# with patterns for sex
ggplot(aes(x = age, y = dx, group = sex)) +
# The ggpattern package creates stacked charts with
# geom_col_pattern different patterns based on the sex
# variable
ggpattern::geom_col_pattern(aes(pattern=sex),
position="stack",
fill= 'white',
colour= 'black') +
# Again, set the title and names for x and y axes
labs(title = "Lung Cancer Deaths by Age in Germany",
x = "Age",
y = "Deaths")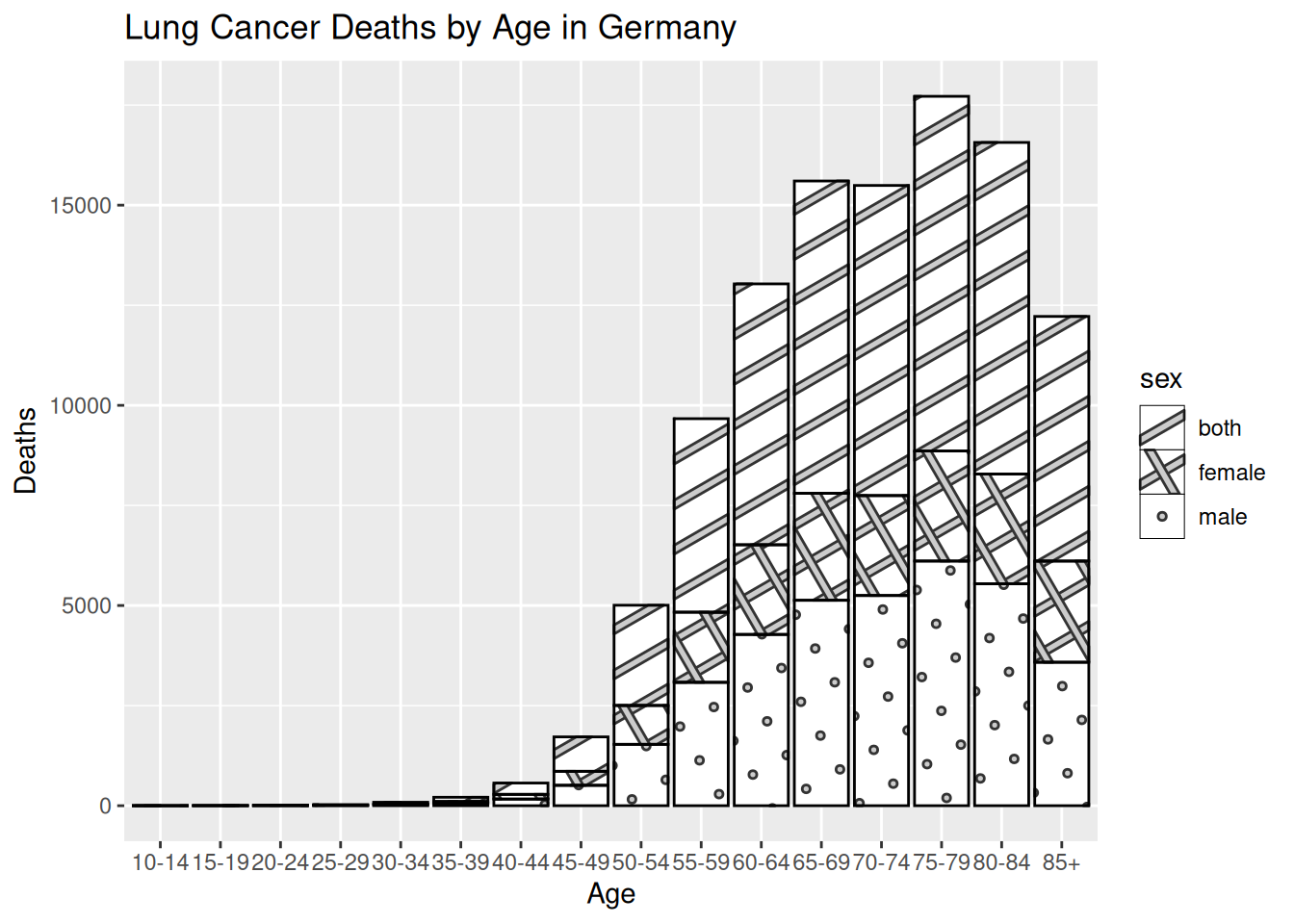
Things to do:
Create a bar chart with
geom_bar.Use faceting to separate the categories with
facet_grid.Angle the x-axis category labels with
hjust.Set the y-axis origin to 0.
# Create a barplot
barplot <- hmsidwR::germany_lungc |>
# This time, the aesthetics are age and number of deaths with
# patterns for sex
ggplot(aes(x = age, y = dx)) +
# The geom_bar argument is used for bar charts
geom_bar(fill = "white",
# stat = "identity" tells R to calculate the sum of
# the y variable, grouped by the x variable
stat = "identity",
# Add a black outline
colour = "black") +
# Set the origin for the y-axis at 0
scale_y_continuous(expand = c(0, 0)) +
# Again, set the title and names for x and y axes
labs(title = "Lung Cancer Deaths by Age in Germany",
x = "Age",
y = "Deaths") +
# Separate panels for each sex category, and modify the
# fonts and some styling
facet_grid(sex ~ .) +
theme_classic() +
theme(text = element_text(family = "Lato",
size = 12),
axis.text = element_text(colour = "black"),
axis.text.x = element_text(angle = 45,
hjust = 1))
barplot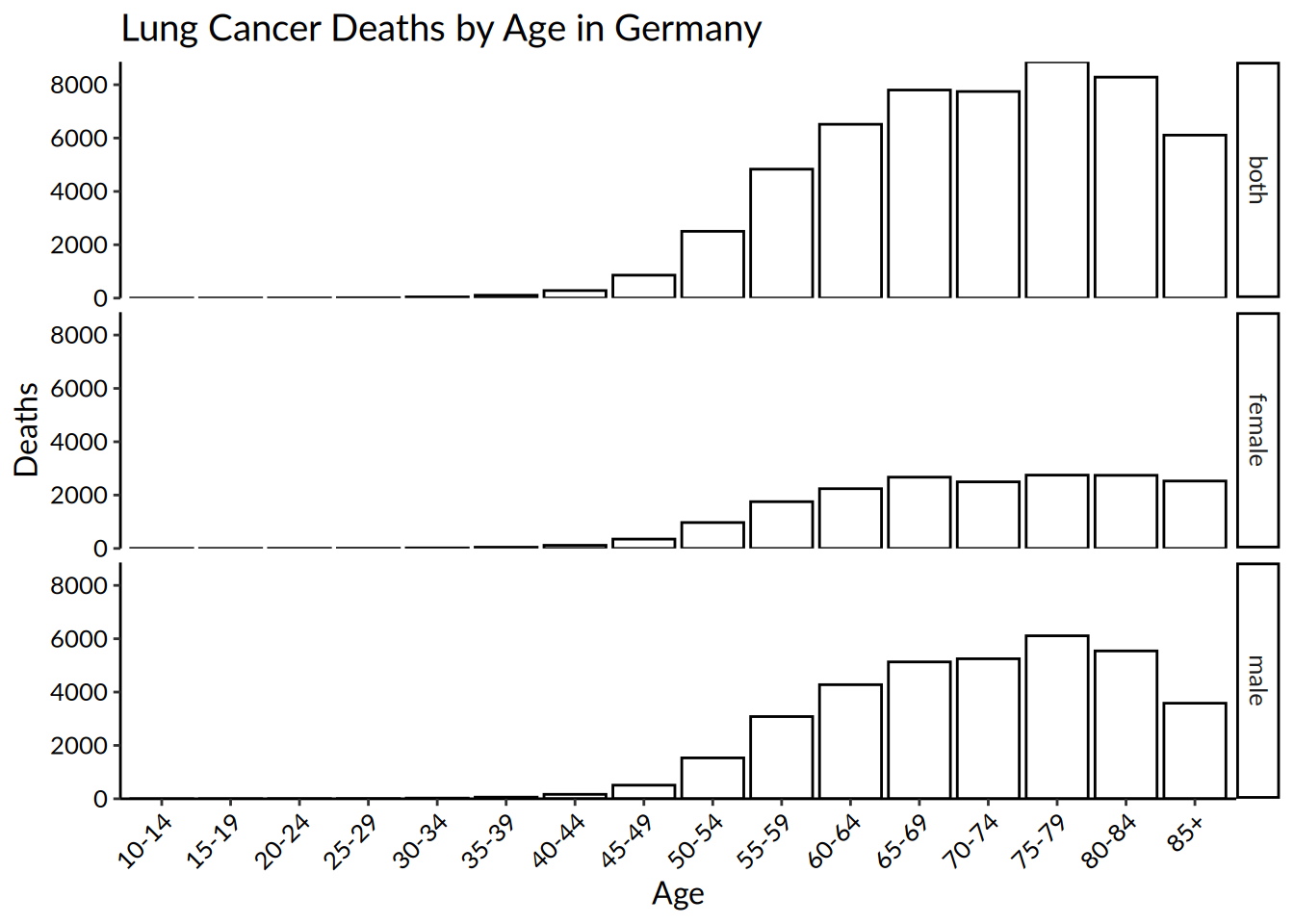
To format the strip text in the facets, use the labeller argument inside facet_grid:
# Put the categories in a list
sex_categories <- list(
"both" = "Both",
"female" = "Female",
"male" = "Male"
)
# Create a function to store the new names
sex_labeller <- function(variable,value){
return(sex_categories[value])
}
# Add the names to the strip panels
barplot +
facet_grid(~ sex, labeller = sex_labeller) +
theme_classic() +
theme(text = element_text(size = 10),
axis.text = element_text(colour = "black"),
axis.text.x = element_text(angle = 45,
hjust = 1))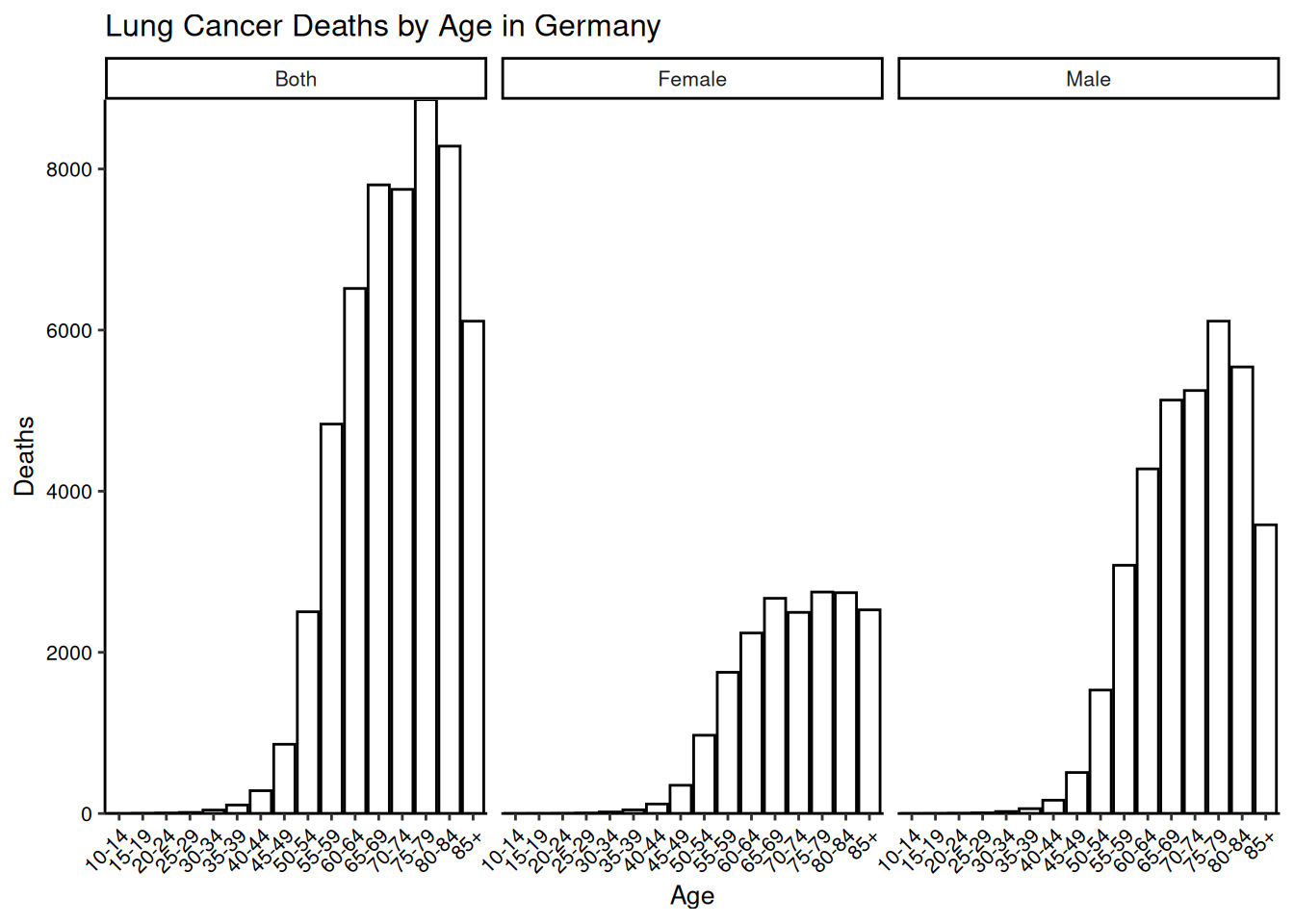
Line plot
lineplot <- hmsidwR::germany_lungc |>
# Variables are age, deaths, grouped by sex
ggplot(aes(x = age, y = dx, group = sex)) +
# geom_line creates the lines, varying the type of line by sex
geom_line(aes(linetype = sex)) +
# Add points with geom_point
geom_point() +
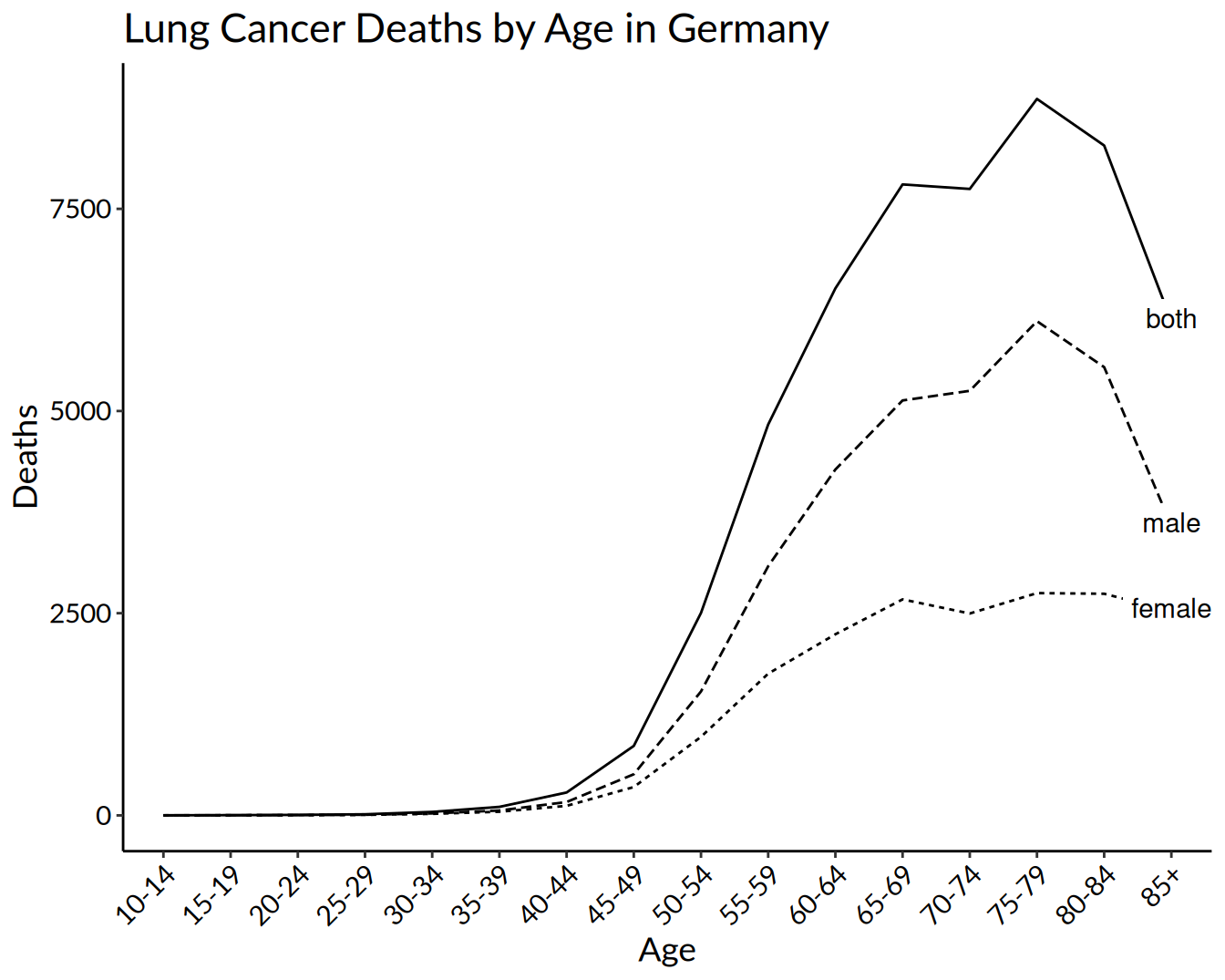
labs(title = "Lung Cancer Deaths by Age in Germany",
x = "Age",
y = "Deaths")
lineplot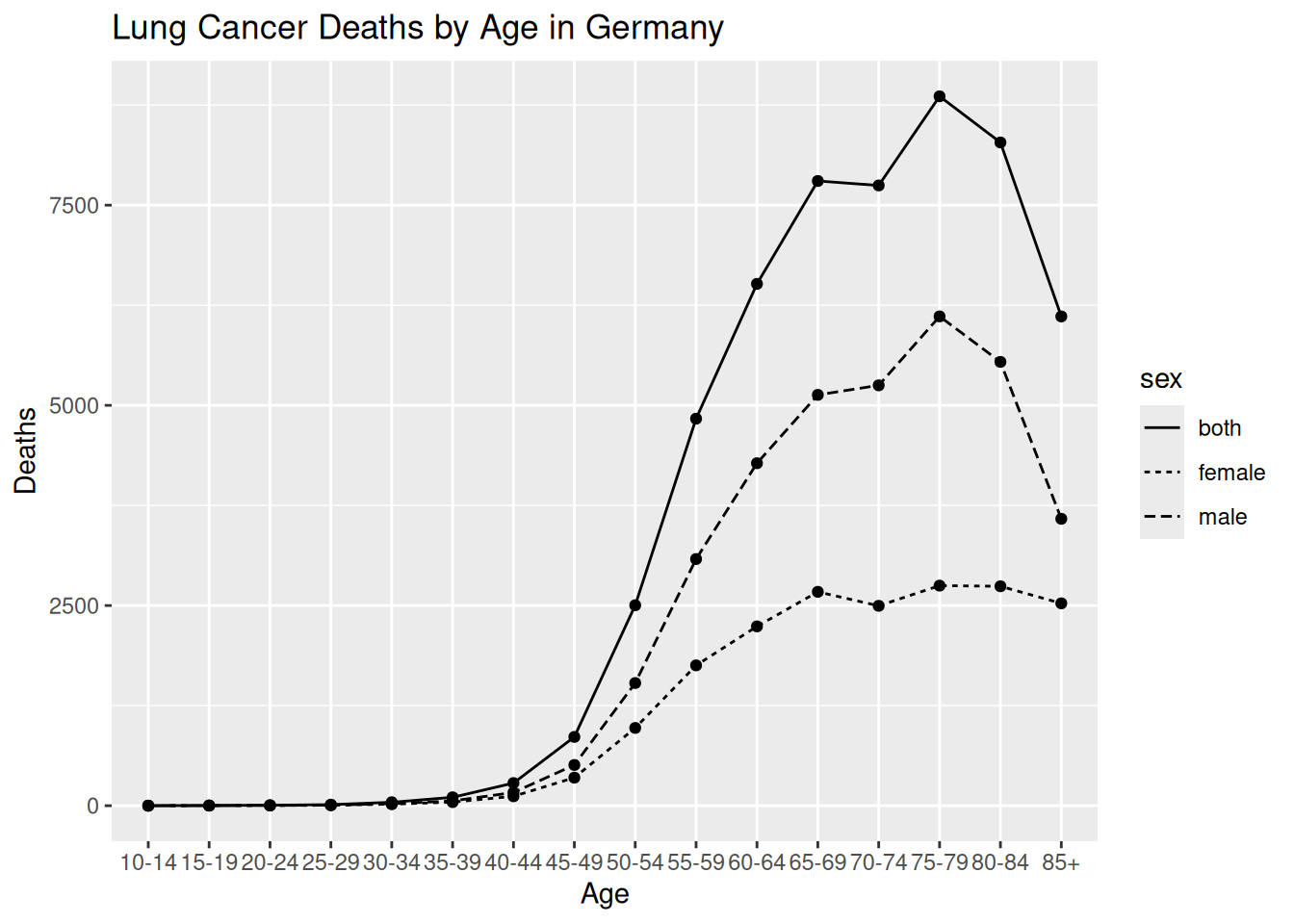
Lineplot
Things to do:
Remove the legend and add labels to the ends of the lines with package
{ggtext}.Plain background and similar formatting as before.
library(ggtext)
# Subset the data to create labels based on the oldest age
# category to put the labels at the end of the line
dat_label <- subset(hmsidwR::germany_lungc, age == max(age))
hmsidwR::germany_lungc |>
# Same variables as before
ggplot(aes(x = age, y = dx, group = sex)) +
# geom_line creates the lines, varying the type of line by
# sex; remove the legend
geom_line(aes(linetype = sex),
show.legend = FALSE) +
# Use the subsetted data created to add the labels
ggtext::geom_richtext(data = dat_label,
label.color = NA,
aes(label = sex)) +
labs(title = "Lung Cancer Deaths by Age in Germany",
x = "Age",
y = "Deaths") +
theme_classic() +
theme(text = element_text(family = "Lato",
size = 14),
axis.text = element_text(colour = "black"),
axis.text.x = element_text(angle = 45,
hjust = 1))