6.3.3 Example: Epidemic Y
6.3.3.1 INLA: an empirical Bayes approach to GAMs
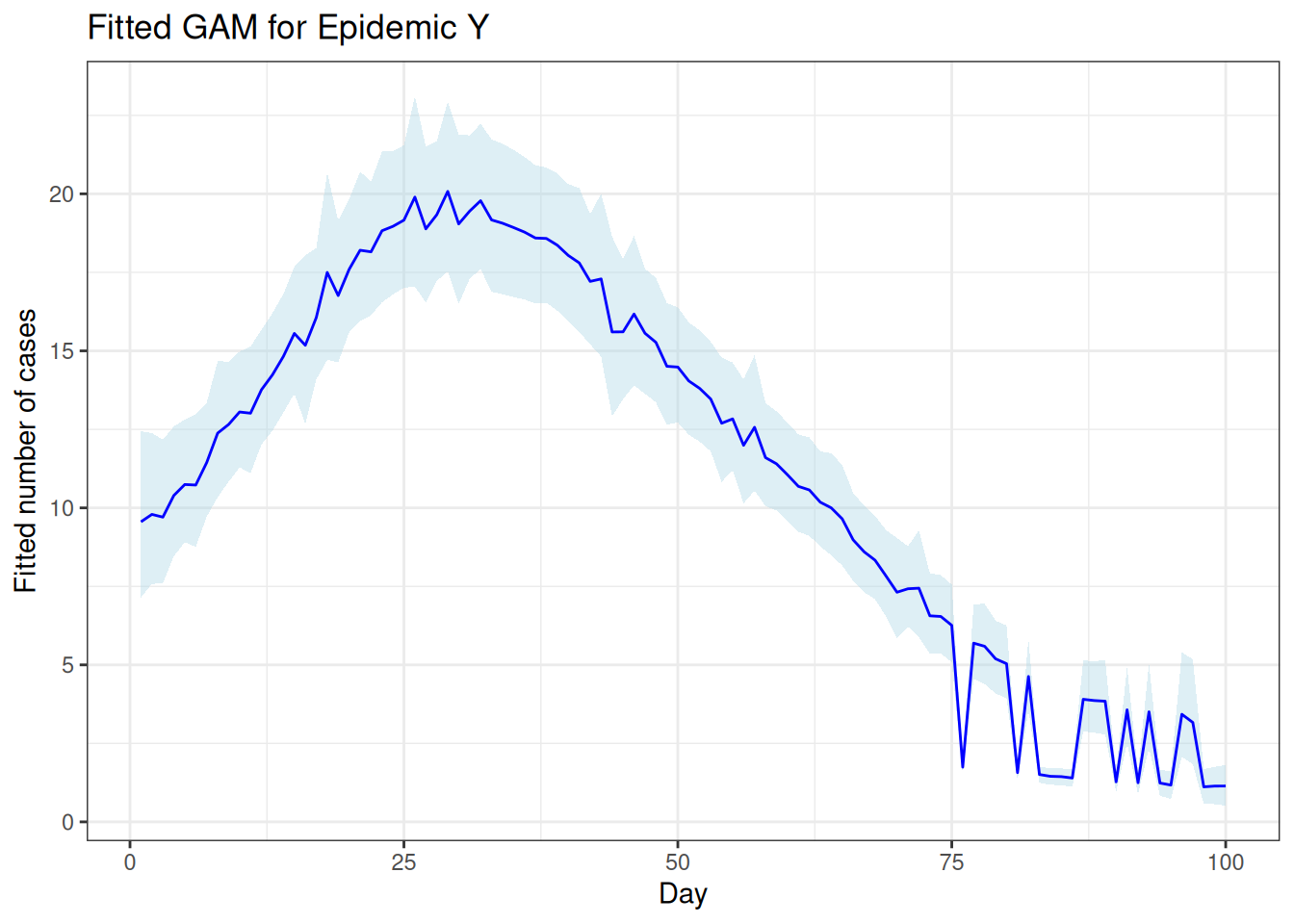
Scenario: epidemic over 100 days based on temperature level and number of cases simulated with a random poisson distribution.
Model: INLA - integrated nested Laplace approximation model.
A deterministic algorithm for Bayesian inference.
Useful for fitting complex models (GAMs).
Alternative to Markov Chain Monte Carlo (MCMC).
Additional reading that may be useful: the R-INLA website, with tutorials and examples and a gentle INLA tutorial.
Install the INLA package (might take a while…)
Create the sample data.
set.seed(123)
epidemic_data <- data.frame(
day = 1:100,
temperature = rnorm(100, mean = 20, sd = 5),
cases = rpois(100,
lambda = 10 + sin(1:100 / 20) * 10 + rnorm(100,
sd = 5)))
# View the first few rows of data
head(epidemic_data)## day temperature cases
## 1 1 17.19762 13
## 2 2 18.84911 8
## 3 3 27.79354 9
## 4 4 20.35254 11
## 5 5 20.64644 6
## 6 6 28.57532 14- Model the number of cases as a function of time and temperature
- non-linear effect of time using a random walk model (model = “rw2”)
- linear effect of temperature
# Define the model formula
formula <- cases ~ f(day, model = "rw2") + temperature
# Fit the model using INLA
result <- inla(formula,
family = "poisson",
data = epidemic_data)Check the results:
## mean sd 0.025quant 0.5quant 0.975quant
## (Intercept) 2.389329032 0.144602391 2.10579390 2.389320218 2.672915611
## temperature -0.004819585 0.006762735 -0.01808339 -0.004818755 0.008439435
## mode kld
## (Intercept) 2.389320552 3.237757e-11
## temperature -0.004818768 4.003960e-11- kld refers to Kullback and Leibler (1951).
The predicted number of cases per day can be extracted and plotted:
time_effect <- result$summary.random$day
fitted_values <- result$summary.fitted.values
# Creating a data frame for plotting
plot_data <- data.frame(Day = epidemic_data$day,
FittedCases = fitted_values$mean,
Lower = fitted_values$`0.025quant`,
Upper = fitted_values$`0.975quant`)