6.3.1 Example: Cholera
Deaths due to cholera over 12 months in 1849 (from the {HistData} package):
library(tidyverse)
library(HistData)
cholera <- HistData::CholeraDeaths1849 %>%
filter(cause_of_death == "Cholera") %>%
select(date, deaths)
cholera %>% head()## # A tibble: 6 × 2
## date deaths
## <date> <dbl>
## 1 1849-01-01 13
## 2 1849-01-02 19
## 3 1849-01-03 28
## 4 1849-01-04 24
## 5 1849-01-05 23
## 6 1849-01-06 39Response (\(y\)) = deaths.
Predictor (\(x\)) = date.
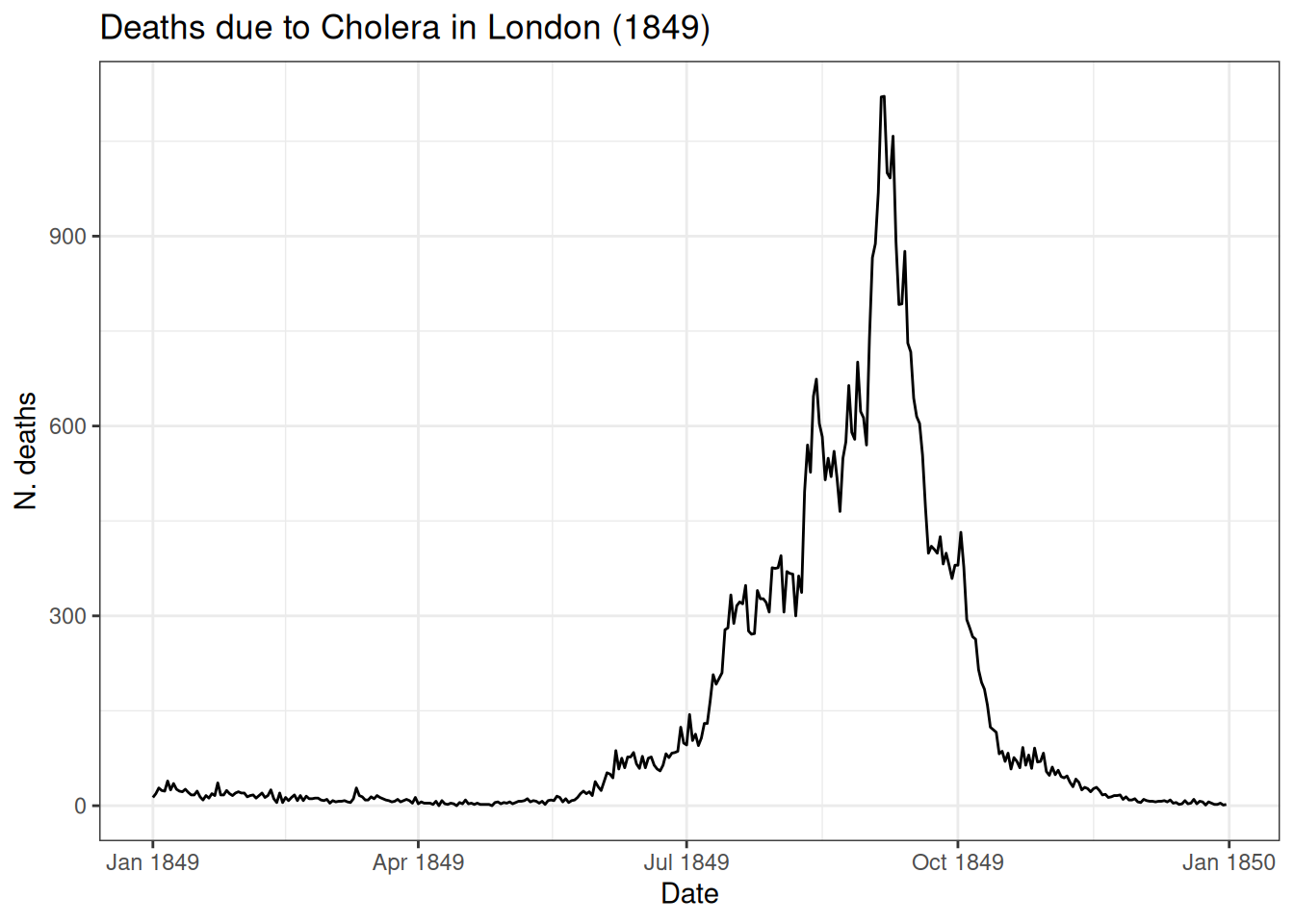
How to model this?
Review the relationship between response and predictor:
Equation of a line: \(y = \beta_0 + \beta_1x\) or (\(y=mx+b\))
\(\beta_0\) - intercept, or value of \(y\) when \(x = 0\)
\(\beta_1\) - average change in \(y\) for each unit increase in \(x\)
\(x\) - value of the predictor
Often there are multiple predictors, adding more complexity to a model.
Modelling estimates these values
Linear models and Locally estimated scatter plot smoothing (LOESS)
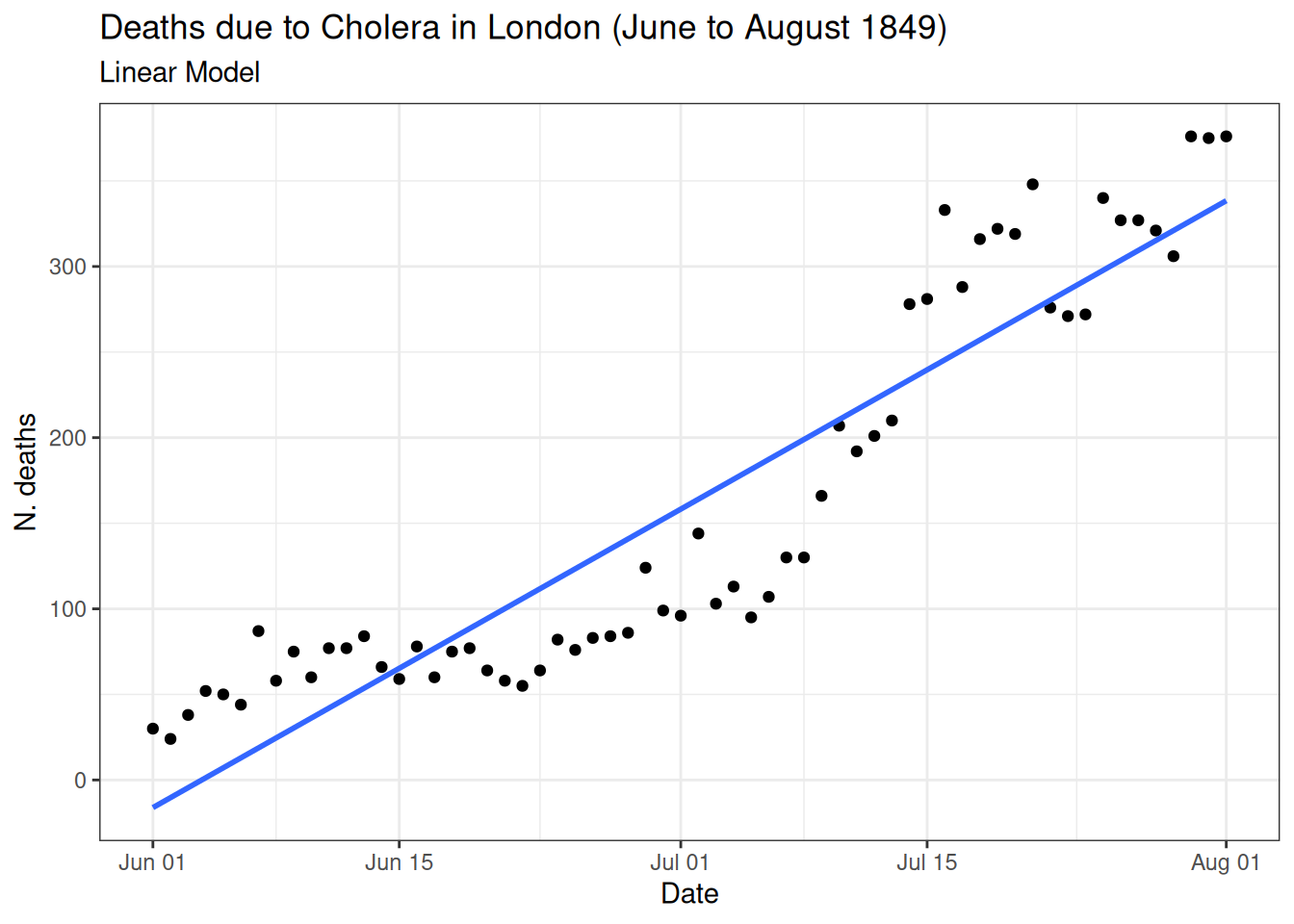
Linear model
- Assumes linear relationship.
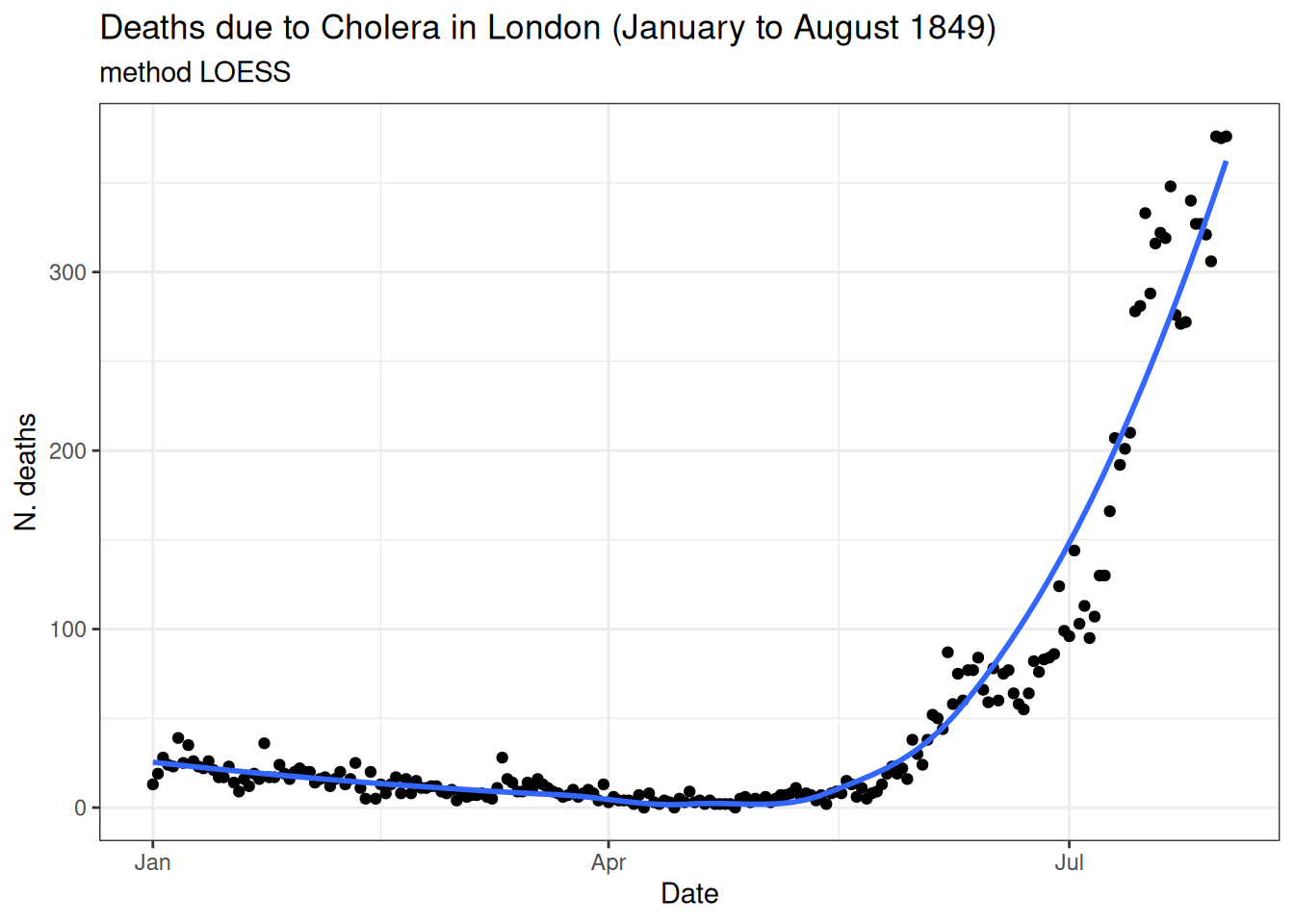
Locally estimated scatter plot smoothing (LOESS)
- No underlying assumptions made about the structure of the data.
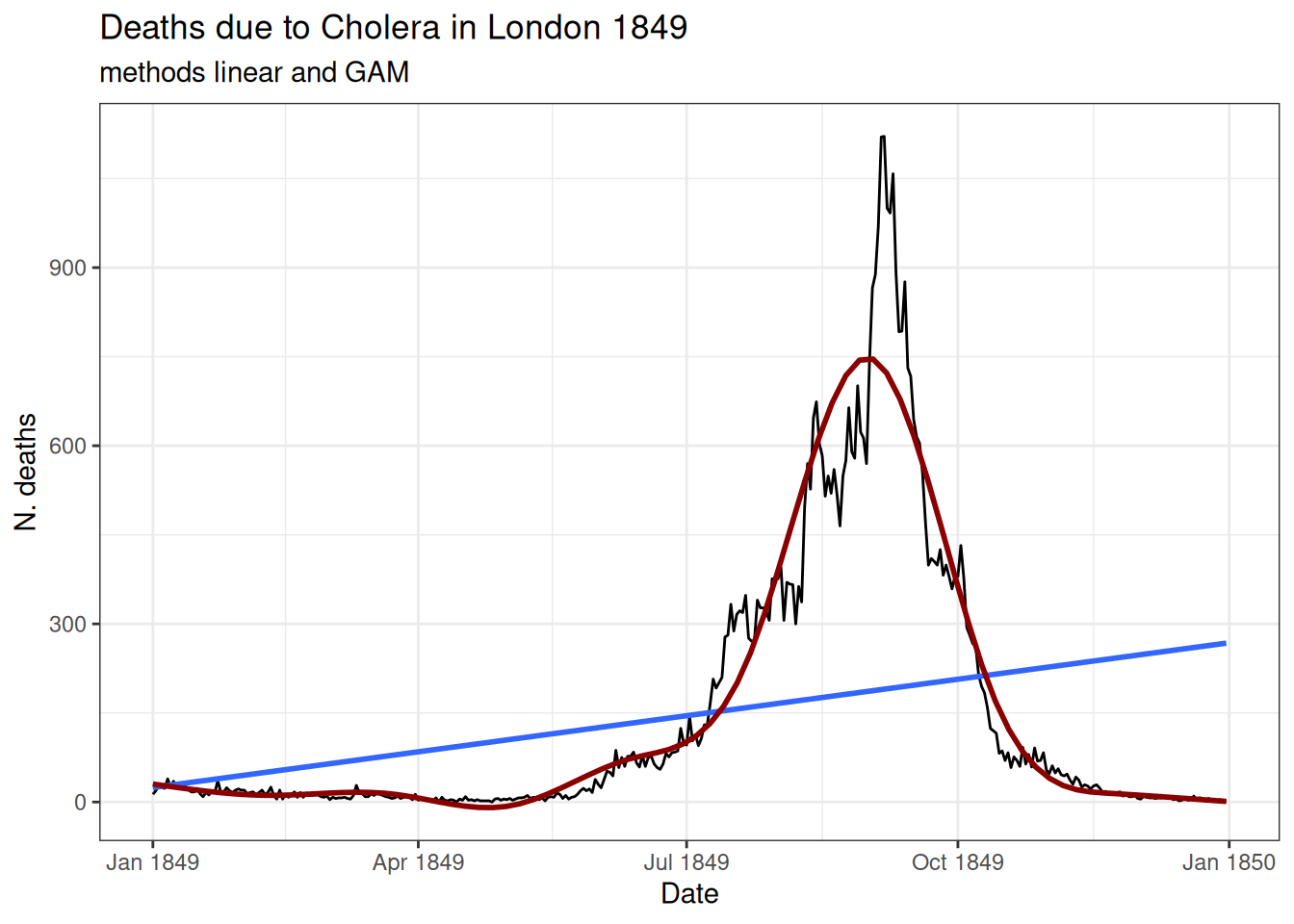
Comparing linear and non-linear models
Plotting a linear model against a generalised additive model (GAM):
GAM - Generalised Additive Model
What are they?
- Flexible extensions of linear models.
- Useful for modelling complex relationships between response (\(y\)) and predictor(s) (\(x_1,...x_n\)).
\(g(\mu) = \beta_0 + f(x)\)
- Predictors are transformed through each function \(x \sim f(x)\).
- Link function \(g(\mu)\) is chosen based on response distribution.
How to do this in R?
# Load the mgcv package
library(mgcv)
# Use the days instead of the full date
cholera$days <- row_number(cholera)
# Look at the first six rows
head(cholera)## # A tibble: 6 × 3
## date deaths days
## <date> <dbl> <int>
## 1 1849-01-01 13 1
## 2 1849-01-02 19 2
## 3 1849-01-03 28 3
## 4 1849-01-04 24 4
## 5 1849-01-05 23 5
## 6 1849-01-06 39 6# Fit a GAM using the gam() function
gam_model <- gam(deaths ~ s(days), data = cholera)
# Print the summary of the GAM model
summary(gam_model)##
## Family: gaussian
## Link function: identity
##
## Formula:
## deaths ~ s(days)
##
## Parametric coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 146.008 3.583 40.74 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Approximate significance of smooth terms:
## edf Ref.df F p-value
## s(days) 8.969 9 431.4 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## R-sq.(adj) = 0.914 Deviance explained = 91.6%
## GCV = 4818.7 Scale est. = 4687 n = 365Key points:
- Intercept is modelled as a parametric term.
- Days is a smooth term: s(days).
- Effective degrees of freedom (EDF) is ~9, indicating a wiggly (non-linear) relationship to deaths.
- Adjusted R-squared value indicates the model explains approx. 91.4% of variance in \(y\).
- Deviance value of 91.6% indicates a good fit.
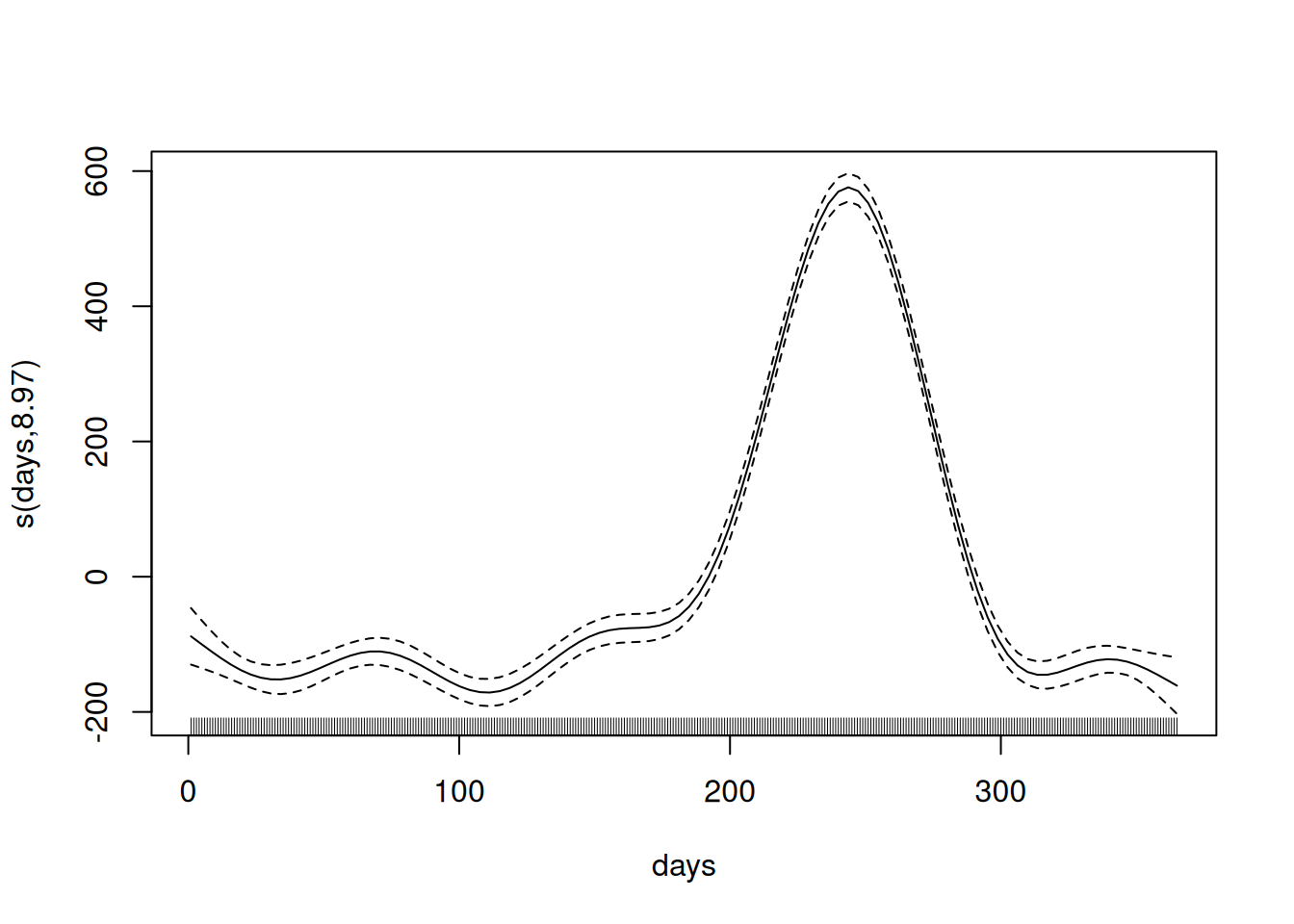
However, the model is very simple with only one predictor. In reality there are likely to be many other factors that influence number of deaths.