12.3 Data-masking
Data-masking functions allow you to use variables in the “current” data frame without any extra syntax. It’s used in many dplyr functions like arrange(), filter(), group_by(), mutate(), and summarise(), and in ggplot2’s aes(). Data-masking is useful because it lets you use data-variables without any additional syntax.
12.3.1 Getting Started
This is a call to filter() which uses a data-variable (carat) and an env-variable (min):
## # A tibble: 17,502 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 1.17 Very Good J I1 60.2 61 2774 6.83 6.9 4.13
## 2 1.01 Premium F I1 61.8 60 2781 6.39 6.36 3.94
## 3 1.01 Fair E I1 64.5 58 2788 6.29 6.21 4.03
## 4 1.01 Premium H SI2 62.7 59 2788 6.31 6.22 3.93
## 5 1.05 Very Good J SI2 63.2 56 2789 6.49 6.45 4.09
## 6 1.05 Fair J SI2 65.8 59 2789 6.41 6.27 4.18
## 7 1.01 Fair E SI2 67.4 60 2797 6.19 6.05 4.13
## 8 1.04 Premium G I1 62.2 58 2801 6.46 6.41 4
## 9 1.2 Fair F I1 64.6 56 2809 6.73 6.66 4.33
## 10 1.02 Premium G I1 60.3 58 2815 6.55 6.5 3.94
## # ℹ 17,492 more rowsThis is its base R equivalent:
## # A tibble: 17,502 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 1.17 Very Good J I1 60.2 61 2774 6.83 6.9 4.13
## 2 1.01 Premium F I1 61.8 60 2781 6.39 6.36 3.94
## 3 1.01 Fair E I1 64.5 58 2788 6.29 6.21 4.03
## 4 1.01 Premium H SI2 62.7 59 2788 6.31 6.22 3.93
## 5 1.05 Very Good J SI2 63.2 56 2789 6.49 6.45 4.09
## 6 1.05 Fair J SI2 65.8 59 2789 6.41 6.27 4.18
## 7 1.01 Fair E SI2 67.4 60 2797 6.19 6.05 4.13
## 8 1.04 Premium G I1 62.2 58 2801 6.46 6.41 4
## 9 1.2 Fair F I1 64.6 56 2809 6.73 6.66 4.33
## 10 1.02 Premium G I1 60.3 58 2815 6.55 6.5 3.94
## # ℹ 17,492 more rowsBase R functions refer to data-variables with $, and you often have to repeat the name of the data frame multiple times, making it clear what is a data-variable and what is an env-variable.
It also makes it straightforward to use indirection because you can store the name of the data-variable in an env-variable, and then switch from $ to [[:
## # A tibble: 17,502 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 1.17 Very Good J I1 60.2 61 2774 6.83 6.9 4.13
## 2 1.01 Premium F I1 61.8 60 2781 6.39 6.36 3.94
## 3 1.01 Fair E I1 64.5 58 2788 6.29 6.21 4.03
## 4 1.01 Premium H SI2 62.7 59 2788 6.31 6.22 3.93
## 5 1.05 Very Good J SI2 63.2 56 2789 6.49 6.45 4.09
## 6 1.05 Fair J SI2 65.8 59 2789 6.41 6.27 4.18
## 7 1.01 Fair E SI2 67.4 60 2797 6.19 6.05 4.13
## 8 1.04 Premium G I1 62.2 58 2801 6.46 6.41 4
## 9 1.2 Fair F I1 64.6 56 2809 6.73 6.66 4.33
## 10 1.02 Premium G I1 60.3 58 2815 6.55 6.5 3.94
## # ℹ 17,492 more rowsWe can achieve the same result with tidy evaluation by somehow adding $ back into the picture while data-masking functions by using .data or .env to be explicit about whether you’re talking about a data-variable or an env-variable:
num_vars <- c("carat", "depth", "table", "price", "x", "y", "z")
ui <- fluidPage(
selectInput("var", "Variable", choices = num_vars),
numericInput("min", "Minimum", value = 1),
tableOutput("output")
)
server <- function(input, output, session) {
data <- reactive(diamonds %>% filter(.data[[input$var]] > .env$input$min))
output$output <- renderTable(head(data()))
}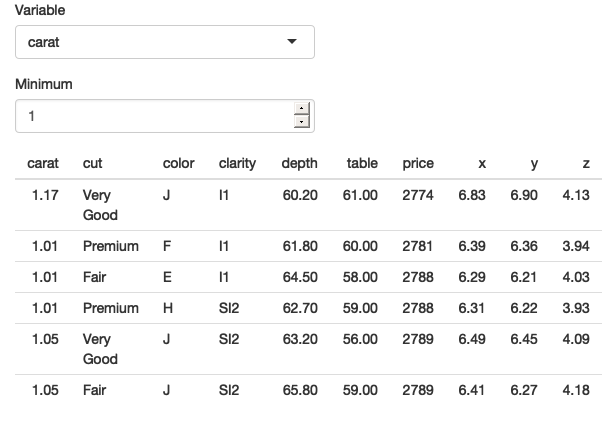
The app works now that we’ve been explicit about .data and .env and [[ vs $. See live at https://hadley.shinyapps.io/ms-tidied-up.
12.3.2 Example: ggplot2
Here we apply this idea to a dynamic plot where we allow the user to create a scatterplot by selecting the variables to appear on the x and y axes. ggforce::position_auto() was used so that geom_point() works regardless of whether the x and y variables are continuous or discrete.
ui <- fluidPage(
selectInput("x", "X variable", choices = names(iris)),
selectInput("y", "Y variable", choices = names(iris)),
plotOutput("plot")
)
server <- function(input, output, session) {
output$plot <- renderPlot({
ggplot(iris, aes(.data[[input$x]], .data[[input$y]])) +
geom_point(position = ggforce::position_auto())
}, res = 96)
}Alternatively, we could allow the user to pick the geom. The following app uses a switch() statement to generate a reactive geom that is later added to the plot.
ui <- fluidPage(
selectInput("x", "X variable", choices = names(iris)),
selectInput("y", "Y variable", choices = names(iris)),
selectInput("geom", "geom", c("point", "smooth", "jitter")),
plotOutput("plot")
)
server <- function(input, output, session) {
plot_geom <- reactive({
switch(input$geom,
point = geom_point(),
smooth = geom_smooth(se = FALSE),
jitter = geom_jitter()
)
})
output$plot <- renderPlot({
ggplot(iris, aes(.data[[input$x]], .data[[input$y]])) +
plot_geom()
}, res = 96)
}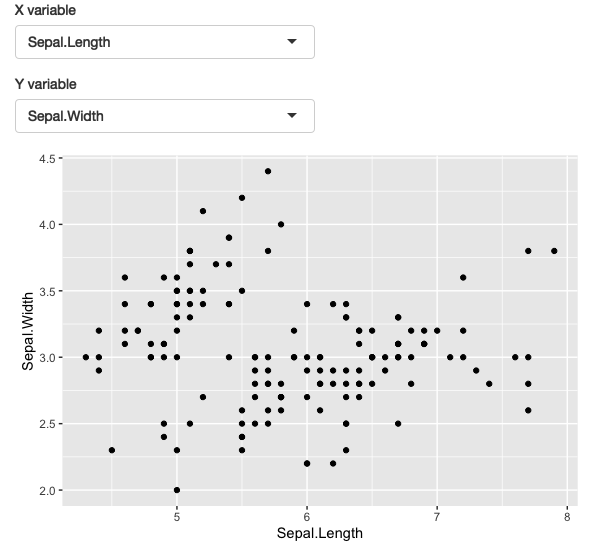
This app allows you to select which variables are plotted on the x and y axes. See live at https://hadley.shinyapps.io/ms-ggplot2.
One of the challenges of programming with user selected variables is that your code has to become more complicated to handle all the cases the user might generate.
12.3.3 Example: dplyr
The same technique also works for dplyr. The following app extends the previous simple example to allow you to choose a variable to filter, a minimum value to select, and a variable to sort by.
ui <- fluidPage(
selectInput("var", "Select variable", choices = names(mtcars)),
sliderInput("min", "Minimum value", 0, min = 0, max = 100),
selectInput("sort", "Sort by", choices = names(mtcars)),
tableOutput("data")
)
server <- function(input, output, session) {
observeEvent(input$var, {
rng <- range(mtcars[[input$var]])
updateSliderInput(
session, "min",
value = rng[[1]],
min = rng[[1]],
max = rng[[2]]
)
})
output$data <- renderTable({
mtcars %>%
filter(.data[[input$var]] > input$min) %>%
arrange(.data[[input$sort]])
})
}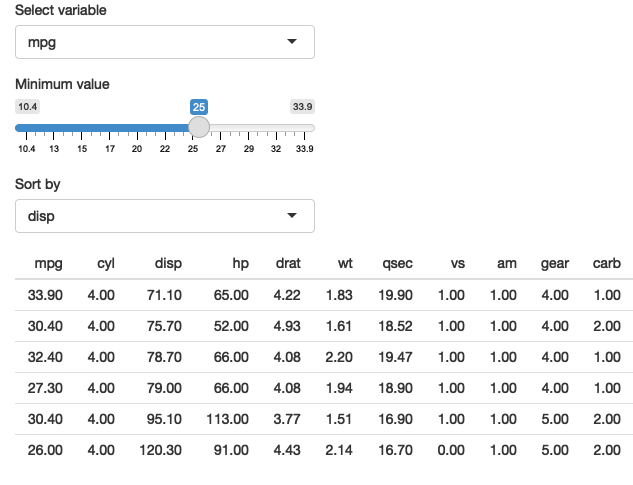
This app that allows you to pick a variable to threshold, and choose how to sort the results. See live at https://hadley.shinyapps.io/ms-dplyr.
Most other problems can be solved by combining .data with your existing programming skills. For example, what if you wanted to conditionally sort in either ascending or descending order?
ui <- fluidPage(
selectInput("var", "Sort by", choices = names(mtcars)),
checkboxInput("desc", "Descending order?"),
tableOutput("data")
)
server <- function(input, output, session) {
sorted <- reactive({
if (input$desc) {
arrange(mtcars, desc(.data[[input$var]]))
} else {
arrange(mtcars, .data[[input$var]])
}
})
output$data <- renderTable(sorted())
}12.3.4 User supplied data
This app allows the user to upload a tsv file, then select a variable and filter by it. It will work for the vast majority of inputs that you might try it with.
ui <- fluidPage(
fileInput("data", "dataset", accept = ".tsv"),
selectInput("var", "var", character()),
numericInput("min", "min", 1, min = 0, step = 1),
tableOutput("output")
)
server <- function(input, output, session) {
data <- reactive({
req(input$data)
vroom::vroom(input$data$datapath)
})
observeEvent(data(), {
updateSelectInput(session, "var", choices = names(data()))
})
observeEvent(input$var, {
val <- data()[[input$var]]
updateNumericInput(session, "min", value = min(val))
})
output$output <- renderTable({
req(input$var)
data() %>%
filter(.data[[input$var]] > input$min) %>%
arrange(.data[[input$var]]) %>%
head(10)
})
}
An app that filter users supplied data, with a surprising failure mode See live at https://hadley.shinyapps.io/ms-user-supplied.
It’ll work with the vast majority of data frames. However, if the data frame contains a variable called input, we get an error message because filter() is attempting to evaluate df$input$min:
df <- data.frame(x = 1, y = 2)
input <- list(var = "x", min = 0)
df %>% filter(.data[[input$var]] > input$min)This problem is due to the ambiguity of data-variables and env-variables, and because data-masking prefers to use a data-variable if both are available. We can resolve the problem by using .env to tell filter() only look for min in the env-variables:
You only need to worry about this problem when working with user supplied data; when working with your own data, you can ensure the names of your data-variables don’t clash with the names of your env-variables.
12.3.5 Why not use base R?
You might wonder if you’re better off without filter(), and if instead you should use the equivalent base R code:
That’s a totally legitimate position, as long as you’re aware of the work that filter() does for you so you can generate the equivalent base R code. In this case:
You’ll need
drop = FALSEif df only contains a single column (otherwise you’ll get a vector instead of a data frame).You’ll need to use
which()or similar to drop any missing values.You can’t do group-wise filtering (e.g.
df %>% group_by(g) %>% filter(n() == 1)).
In general, if you’re using dplyr for very simple cases, you might find it easier to use base R functions that don’t use data-masking. However, in my opinion, one of the advantages of the tidyverse is the careful thought that has been applied to edge cases so that functions work more consistently.