8.5 Visualizing Missing Information
Load scat dataset
data(scat)
scat %>%
glimpse()## Rows: 110
## Columns: 19
## $ Species <fct> coyote, coyote, bobcat, coyote, coyote, coyote, bobcat, bobc…
## $ Month <fct> January, January, January, January, January, January, Januar…
## $ Year <int> 2012, 2012, 2012, 2012, 2012, 2012, 2012, 2012, 2012, 2012, …
## $ Site <fct> YOLA, YOLA, YOLA, YOLA, YOLA, YOLA, ANNU, ANNU, ANNU, ANNU, …
## $ Location <fct> edge, edge, middle, middle, edge, edge, off_edge, off_edge, …
## $ Age <int> 5, 3, 3, 5, 5, 5, 1, 3, 5, 5, 3, 1, 3, 3, 1, 5, 5, 5, 5, 3, …
## $ Number <int> 2, 2, 2, 2, 4, 3, 5, 7, 2, 1, 1, 1, 1, 1, 1, 1, 7, 6, 4, 3, …
## $ Length <dbl> 9.5, 14.0, 9.0, 8.5, 8.0, 9.0, 6.0, 5.5, 11.0, 20.5, 8.0, 8.…
## $ Diameter <dbl> 25.7, 25.4, 18.8, 18.1, 20.7, 21.2, 15.7, 21.9, 17.5, 18.0, …
## $ Taper <dbl> 41.9, 37.1, 16.5, 24.7, 20.1, 28.5, 8.2, 19.3, 29.1, 21.4, N…
## $ TI <dbl> 1.63, 1.46, 0.88, 1.36, 0.97, 1.34, 0.52, 0.88, 1.66, 1.19, …
## $ Mass <dbl> 15.89, 17.61, 8.40, 7.40, 25.45, 14.14, 14.82, 26.41, 16.24,…
## $ d13C <dbl> -26.85, -29.62, -28.73, -20.07, -23.24, -29.00, -28.06, -27.…
## $ d15N <dbl> 6.94, 9.87, 8.52, 5.79, 7.01, 8.28, 4.20, 3.89, 7.34, 6.06, …
## $ CN <dbl> 8.50, 11.30, 8.10, 11.50, 10.60, 9.00, 5.40, 5.60, 5.80, 7.7…
## $ ropey <int> 0, 0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 1, 0, 0, 0, 1, 1, 1, 1, 1, …
## $ segmented <int> 0, 0, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 1, …
## $ flat <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 1, 1, 0, 0, 0, 0, 0, …
## $ scrape <int> 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …Skim scat
skimr::skim(scat) %>%
knitr::kable()| skim_type | skim_variable | n_missing | complete_rate | factor.ordered | factor.n_unique | factor.top_counts | numeric.mean | numeric.sd | numeric.p0 | numeric.p25 | numeric.p50 | numeric.p75 | numeric.p100 | numeric.hist |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| factor | Species | 0 | 1.0000000 | FALSE | 3 | bob: 57, coy: 28, gra: 25 | NA | NA | NA | NA | NA | NA | NA | NA |
| factor | Month | 0 | 1.0000000 | FALSE | 9 | Nov: 17, Jan: 16, Apr: 14, Sep: 14 | NA | NA | NA | NA | NA | NA | NA | NA |
| factor | Site | 0 | 1.0000000 | FALSE | 2 | ANN: 92, YOL: 18 | NA | NA | NA | NA | NA | NA | NA | NA |
| factor | Location | 0 | 1.0000000 | FALSE | 3 | mid: 47, edg: 38, off: 25 | NA | NA | NA | NA | NA | NA | NA | NA |
| numeric | Year | 0 | 1.0000000 | NA | NA | NA | 2011.9363636 | 0.7074605 | 2011.00 | 2011.0000 | 2012.000 | 2012.000 | 2013.00 | ▅▁▇▁▃ |
| numeric | Age | 0 | 1.0000000 | NA | NA | NA | 3.3454545 | 1.3709728 | 1.00 | 3.0000 | 3.000 | 5.000 | 5.00 | ▃▁▇▃▆ |
| numeric | Number | 0 | 1.0000000 | NA | NA | NA | 2.6181818 | 1.4270121 | 1.00 | 2.0000 | 2.000 | 3.000 | 7.00 | ▇▃▂▁▁ |
| numeric | Length | 0 | 1.0000000 | NA | NA | NA | 9.2981818 | 3.4372749 | 2.50 | 6.5000 | 9.000 | 11.500 | 20.50 | ▆▇▇▂▁ |
| numeric | Diameter | 6 | 0.9454545 | NA | NA | NA | 18.5586538 | 3.8820126 | 7.80 | 16.0750 | 18.050 | 21.325 | 30.00 | ▁▅▇▅▁ |
| numeric | Taper | 17 | 0.8454545 | NA | NA | NA | 27.4333333 | 15.0551330 | 2.30 | 17.3000 | 25.800 | 37.400 | 91.50 | ▇▇▃▁▁ |
| numeric | TI | 17 | 0.8454545 | NA | NA | NA | 1.6015054 | 1.0061106 | 0.23 | 0.9900 | 1.430 | 1.890 | 8.68 | ▇▂▁▁▁ |
| numeric | Mass | 1 | 0.9909091 | NA | NA | NA | 12.4552294 | 8.8487894 | 0.94 | 5.6600 | 9.750 | 17.610 | 53.70 | ▇▃▂▁▁ |
| numeric | d13C | 2 | 0.9818182 | NA | NA | NA | -26.8601852 | 2.1755519 | -29.85 | -28.0825 | -27.470 | -26.445 | -19.67 | ▇▇▂▂▁ |
| numeric | d15N | 2 | 0.9818182 | NA | NA | NA | 7.4364815 | 3.0164537 | 1.84 | 5.6200 | 6.885 | 8.305 | 18.00 | ▂▇▂▁▁ |
| numeric | CN | 2 | 0.9818182 | NA | NA | NA | 8.3987963 | 3.6622504 | 4.50 | 6.2000 | 7.250 | 8.650 | 23.60 | ▇▂▁▁▁ |
| numeric | ropey | 0 | 1.0000000 | NA | NA | NA | 0.5636364 | 0.4982036 | 0.00 | 0.0000 | 1.000 | 1.000 | 1.00 | ▆▁▁▁▇ |
| numeric | segmented | 0 | 1.0000000 | NA | NA | NA | 0.5636364 | 0.4982036 | 0.00 | 0.0000 | 1.000 | 1.000 | 1.00 | ▆▁▁▁▇ |
| numeric | flat | 0 | 1.0000000 | NA | NA | NA | 0.0545455 | 0.2281302 | 0.00 | 0.0000 | 0.000 | 0.000 | 1.00 | ▇▁▁▁▁ |
| numeric | scrape | 0 | 1.0000000 | NA | NA | NA | 0.0454545 | 0.2092522 | 0.00 | 0.0000 | 0.000 | 0.000 | 1.00 | ▇▁▁▁▁ |
Plot missing values
vis_dat(scat)
scat %>%
plot_missing()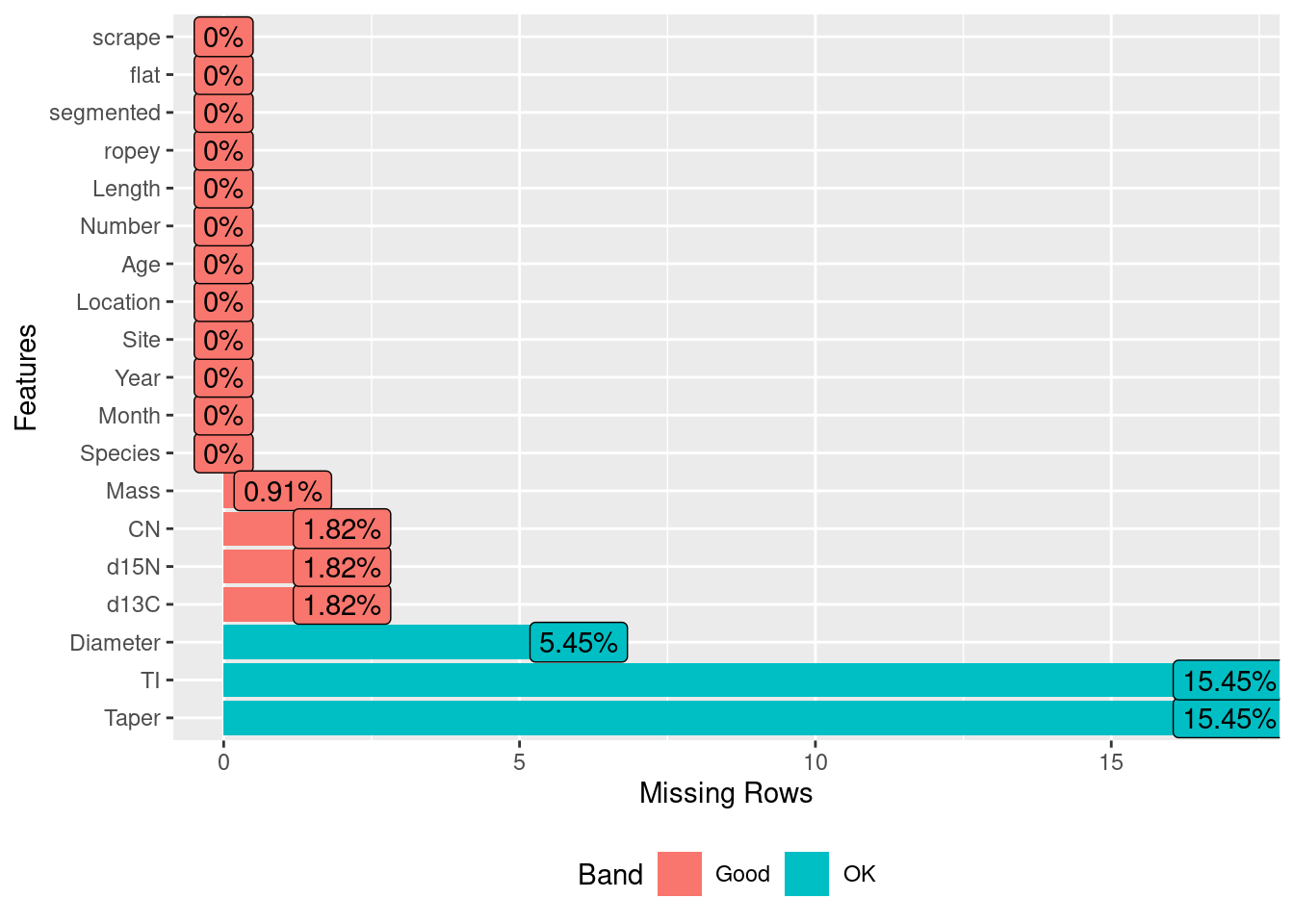
vis_miss(scat)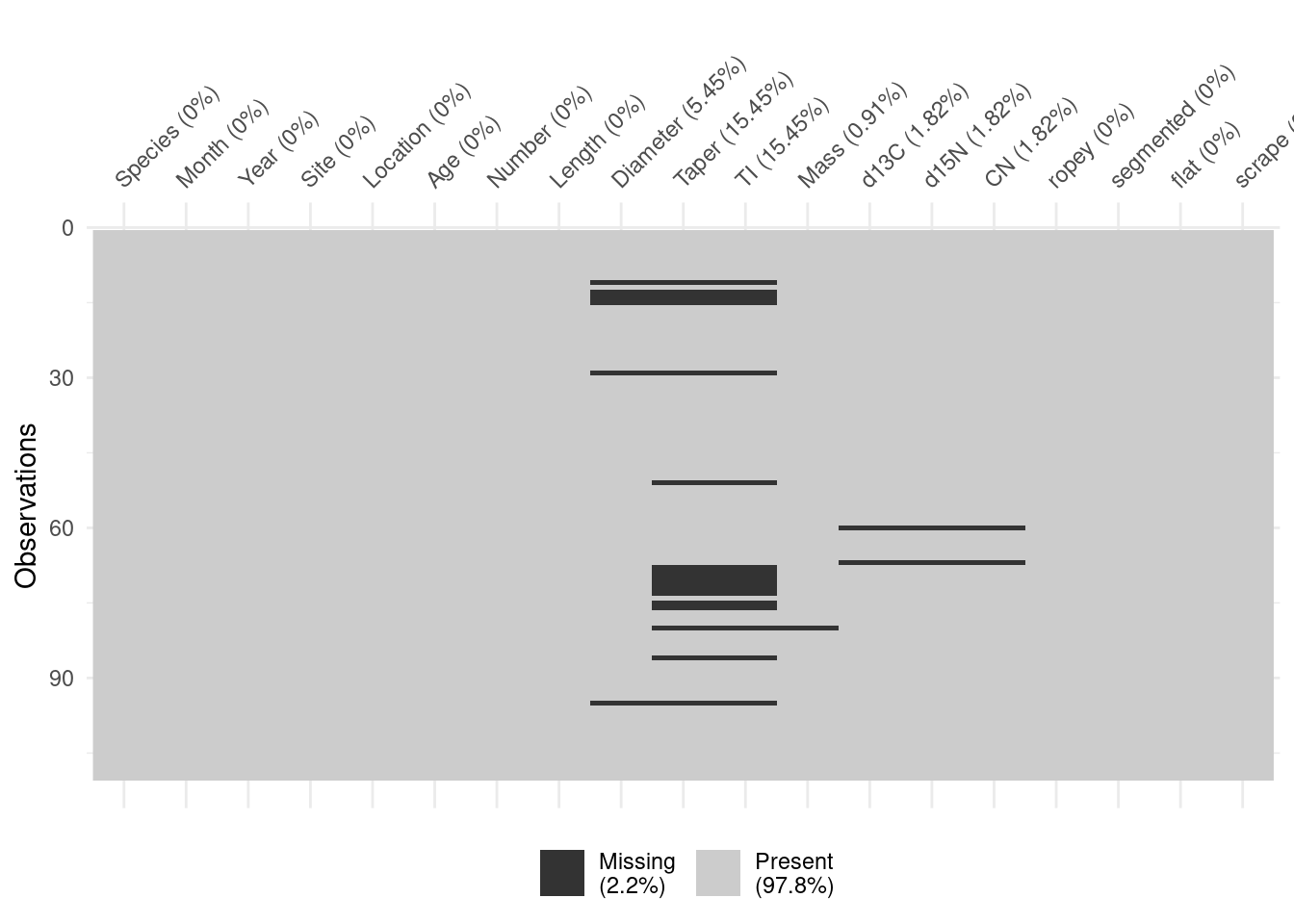
Plot pattern of missingness using an upset plot
gg_miss_upset(scat, nsets = 7)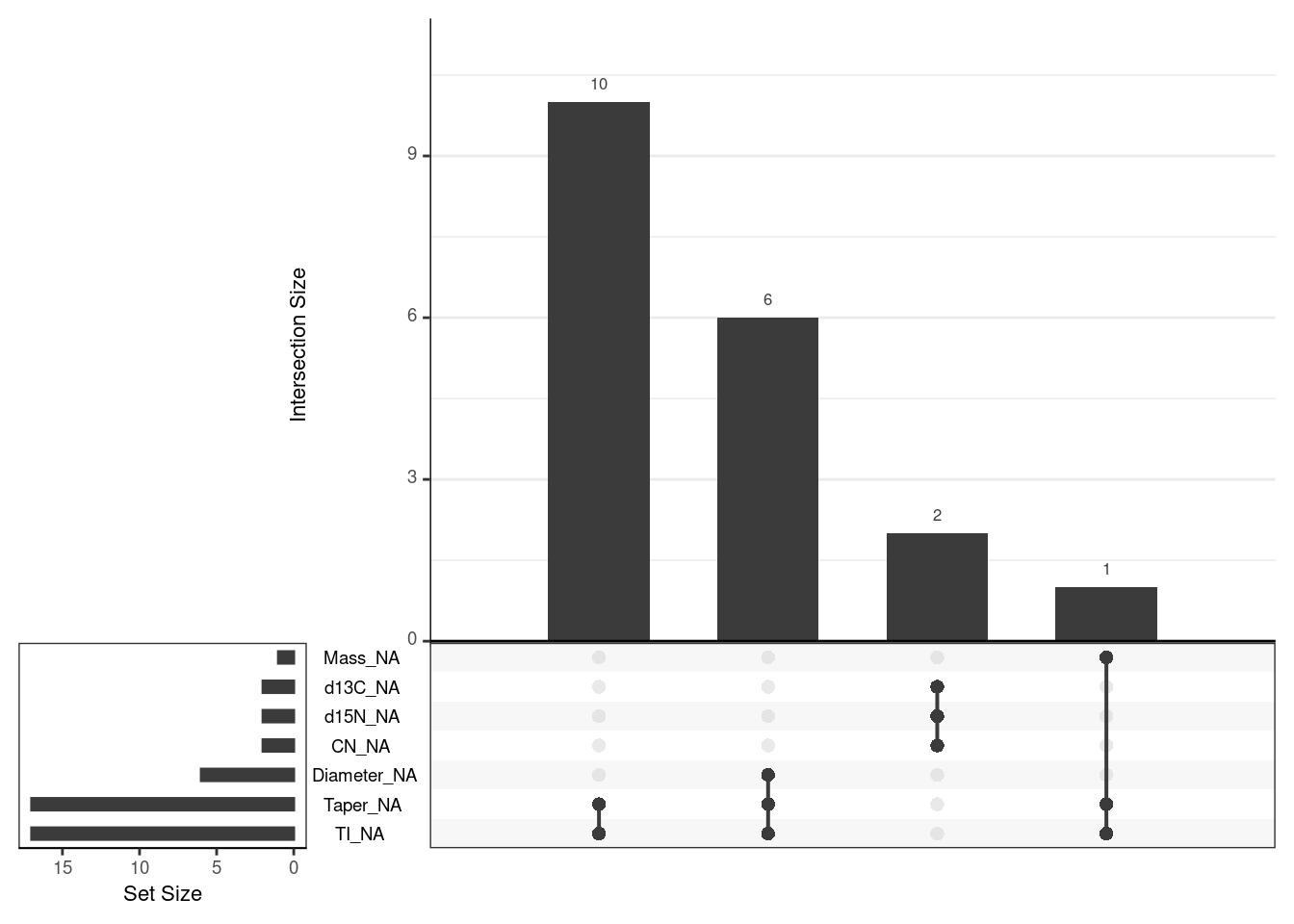
MCAR test
mcar_test(scat)## # A tibble: 1 × 4
## statistic df p.value missing.patterns
## <dbl> <dbl> <dbl> <int>
## 1 169. 65 3.64e-11 5The MCAR hypothesis test result in a p-value < 0.05, indicating that the missing data mechanism is not random.