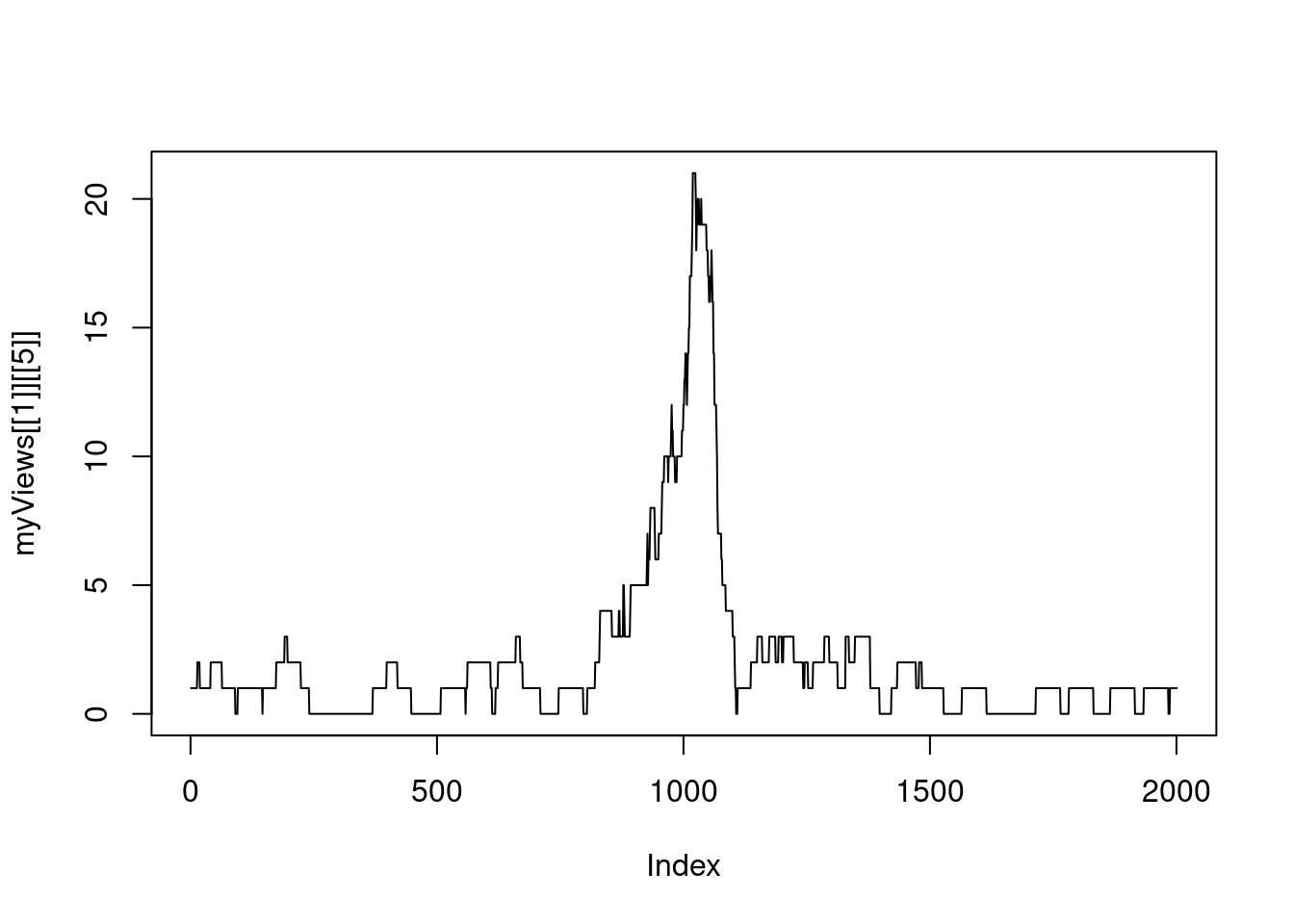
6.3 Rle Vector compression
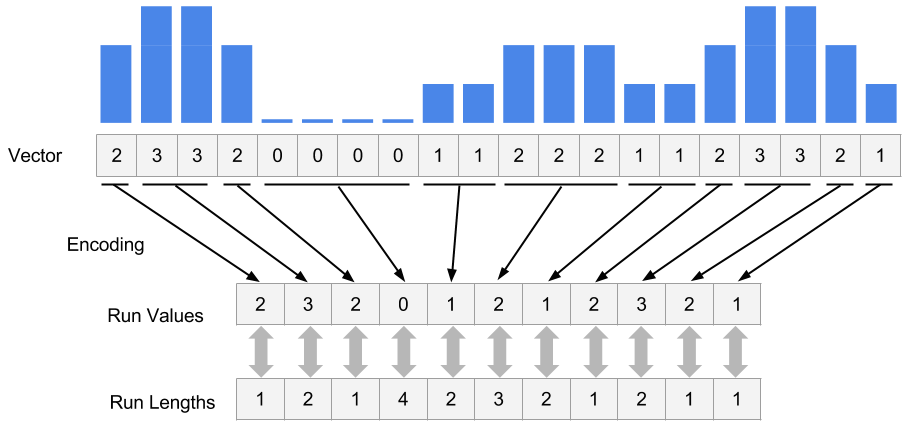
Rle
## RleList of length 24
## $chr1
## integer-Rle of length 249250621 with 1 run
## Lengths: 249250621
## Values : 0
##
## $chr2
## integer-Rle of length 243199373 with 1 run
## Lengths: 243199373
## Values : 0
##
## $chr3
## integer-Rle of length 198022430 with 1 run
## Lengths: 198022430
## Values : 0
##
## $chr4
## integer-Rle of length 191154276 with 1 run
## Lengths: 191154276
## Values : 0
##
## $chr5
## integer-Rle of length 180915260 with 1 run
## Lengths: 180915260
## Values : 0
##
## ...
## <19 more elements>## RleList of length 24
## $chr1
## integer-Rle of length 249250621 with 1 run
## Lengths: 249250621
## Values : 0
##
## $chr2
## integer-Rle of length 243199373 with 1 run
## Lengths: 243199373
## Values : 0
##
## $chr3
## integer-Rle of length 198022430 with 1 run
## Lengths: 198022430
## Values : 0
##
## $chr4
## integer-Rle of length 191154276 with 1 run
## Lengths: 191154276
## Values : 0
##
## $chr5
## integer-Rle of length 180915260 with 1 run
## Lengths: 180915260
## Values : 0
##
## ...
## <19 more elements>library(rtracklayer)
bwFile=system.file("extdata",
"wgEncodeHaibTfbsA549.chr21.bw",
package="compGenomRData")
bw.gr=import(bwFile, which=promoter.gr) # get coverage vectors
bw.gr## GRanges object with 9205 ranges and 1 metadata column:
## seqnames ranges strand | score
## <Rle> <IRanges> <Rle> | <numeric>
## [1] chr21 9825456-9825457 * | 1
## [2] chr21 9825458-9825464 * | 2
## [3] chr21 9825465-9825466 * | 4
## [4] chr21 9825467-9825470 * | 5
## [5] chr21 9825471 * | 6
## ... ... ... ... . ...
## [9201] chr21 48055809-48055856 * | 2
## [9202] chr21 48055857-48055858 * | 1
## [9203] chr21 48055872-48055921 * | 1
## [9204] chr21 48055944-48055993 * | 1
## [9205] chr21 48056069-48056118 * | 1
## -------
## seqinfo: 1 sequence from an unspecified genomecov.bw=coverage(bw.gr,weight = "score") # create Rle List object from GRanges object
cov.bw=import(bwFile, which=promoter.gr,as = "RleList") # or like this
myViews=Views(cov.bw,as(promoter.gr,"IRangesList")) # get subsets of coverage
myViews## RleViewsList object of length 1:
## $chr21
## Views on a 48129895-length Rle subject
##
## views:
## start end width
## [1] 42218039 42220039 2001 [2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [2] 17441841 17443841 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [3] 17565698 17567698 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [4] 30395937 30397937 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [5] 27542138 27544138 2001 [1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 1 1 1 ...]
## [6] 27511708 27513708 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [7] 32930290 32932290 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [8] 27542446 27544446 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [9] 28338439 28340439 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## ... ... ... ... ...
## [370] 47517032 47519032 2001 [1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ...]
## [371] 47648157 47650157 2001 [1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ...]
## [372] 47603373 47605373 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [373] 47647738 47649738 2001 [2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 1 2 ...]
## [374] 47704236 47706236 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [375] 47742785 47744785 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [376] 47881383 47883383 2001 [1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ...]
## [377] 48054506 48056506 2001 [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 ...]
## [378] 48024035 48026035 2001 [1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ...]## [1] 0.2258871 0.3498251 1.2243878 0.4997501 2.0904548 0.6996502## [1] 2 4 12 4 21 6