8.6 Functional enrichment analysis
GO term analysis
library(DESeq2)
library(gProfileR)
library(knitr)
# extract differential expression results
DEresults <- results(dds, contrast = c('group', 'CASE', 'CTRL'))
#remove genes with NA values
DE <- DEresults[!is.na(DEresults$padj),]
#select genes with adjusted p-values below 0.1
DE <- DE[DE$padj < 0.1,]
#select genes with absolute log2 fold change above 1 (two-fold change)
DE <- DE[abs(DE$log2FoldChange) > 1,]
#get the list of genes of interest
genesOfInterest <- rownames(DE)
#calculate enriched GO terms
goResults <- gprofiler(query = genesOfInterest,
organism = 'hsapiens',
src_filter = 'GO',
hier_filtering = 'moderate')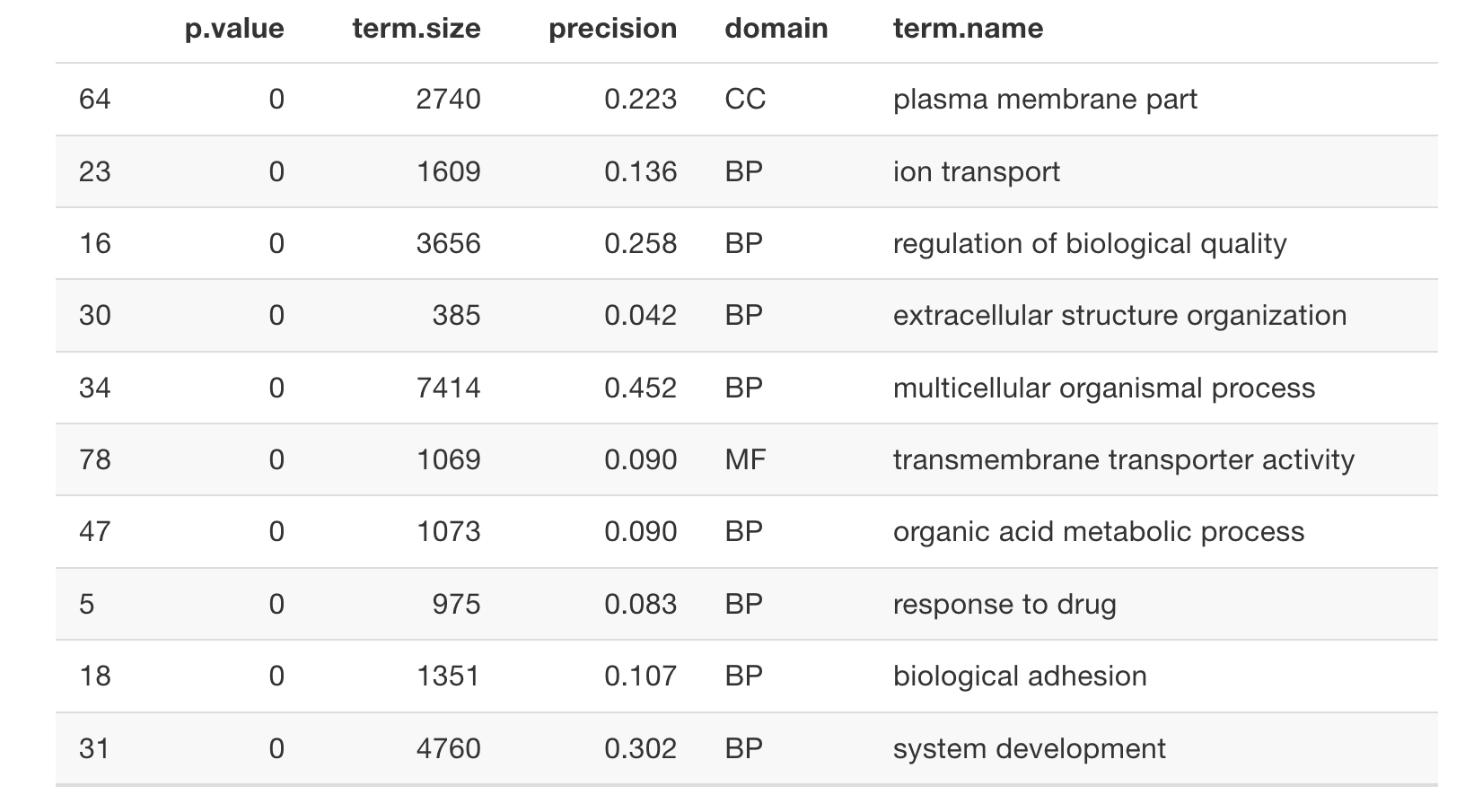
GO_results
Gene set enrichment analysis
… is a valuable exploratory analysis tool that can associate systematic changes to a high-level function rather than individual genes.