5.2 Data pre-processing
Transform –> filter/scale –> handle missing values
#caret for pre-processing
library(caret)
#some of the un-transformed data
boxplot(gexp[,1:50],outline=FALSE,col="cornflowerblue")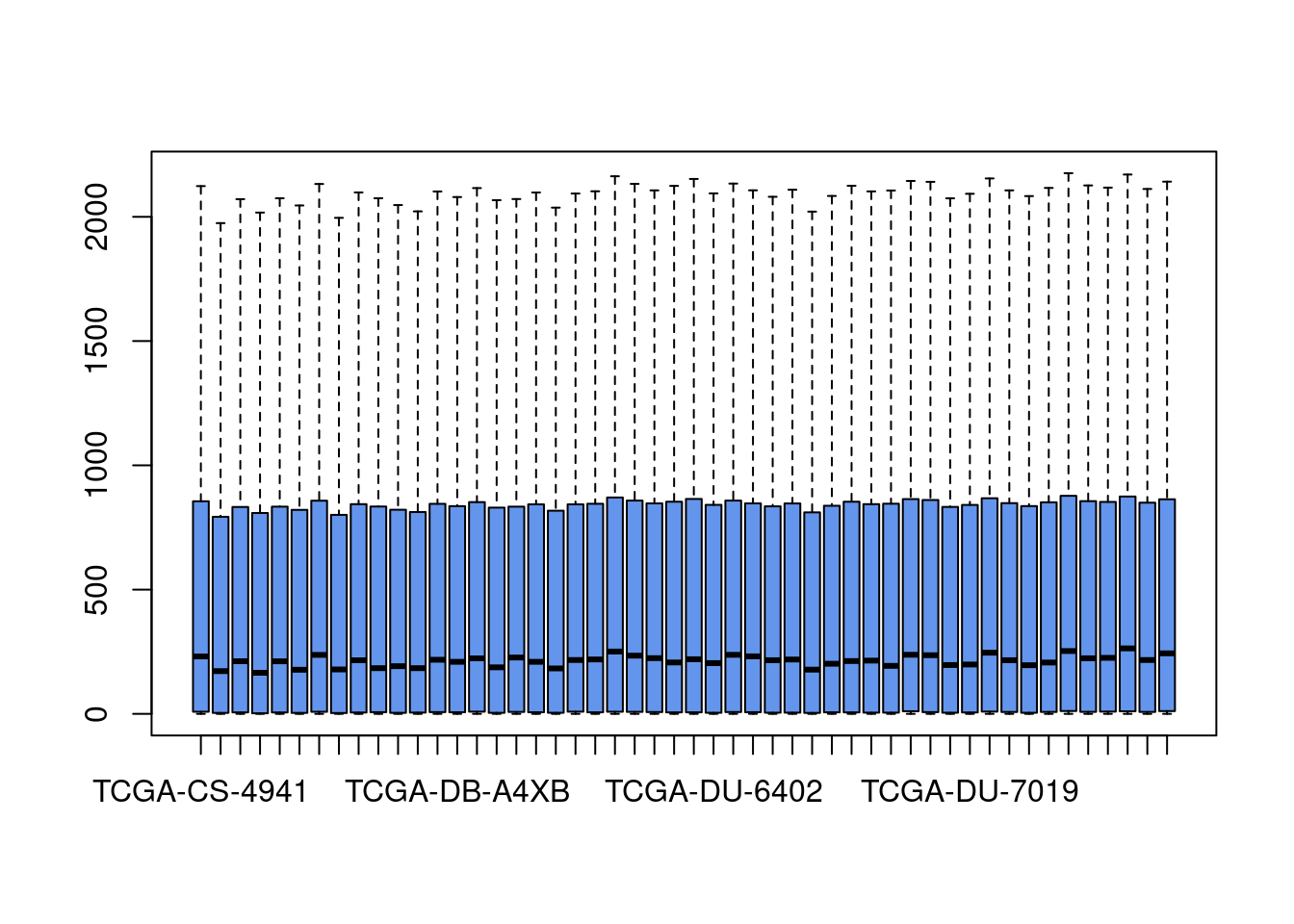
#looks a little funny... let's see what happens when we log transform it.
par(mfrow=c(1,2))
##not transformed
hist(gexp[,5],xlab="gene expression",main="",border="blue4",
col="cornflowerblue")
#log transformed
hist(log10(gexp+1)[,5], xlab="gene expression log scale",main="",
border="blue4",col="cornflowerblue")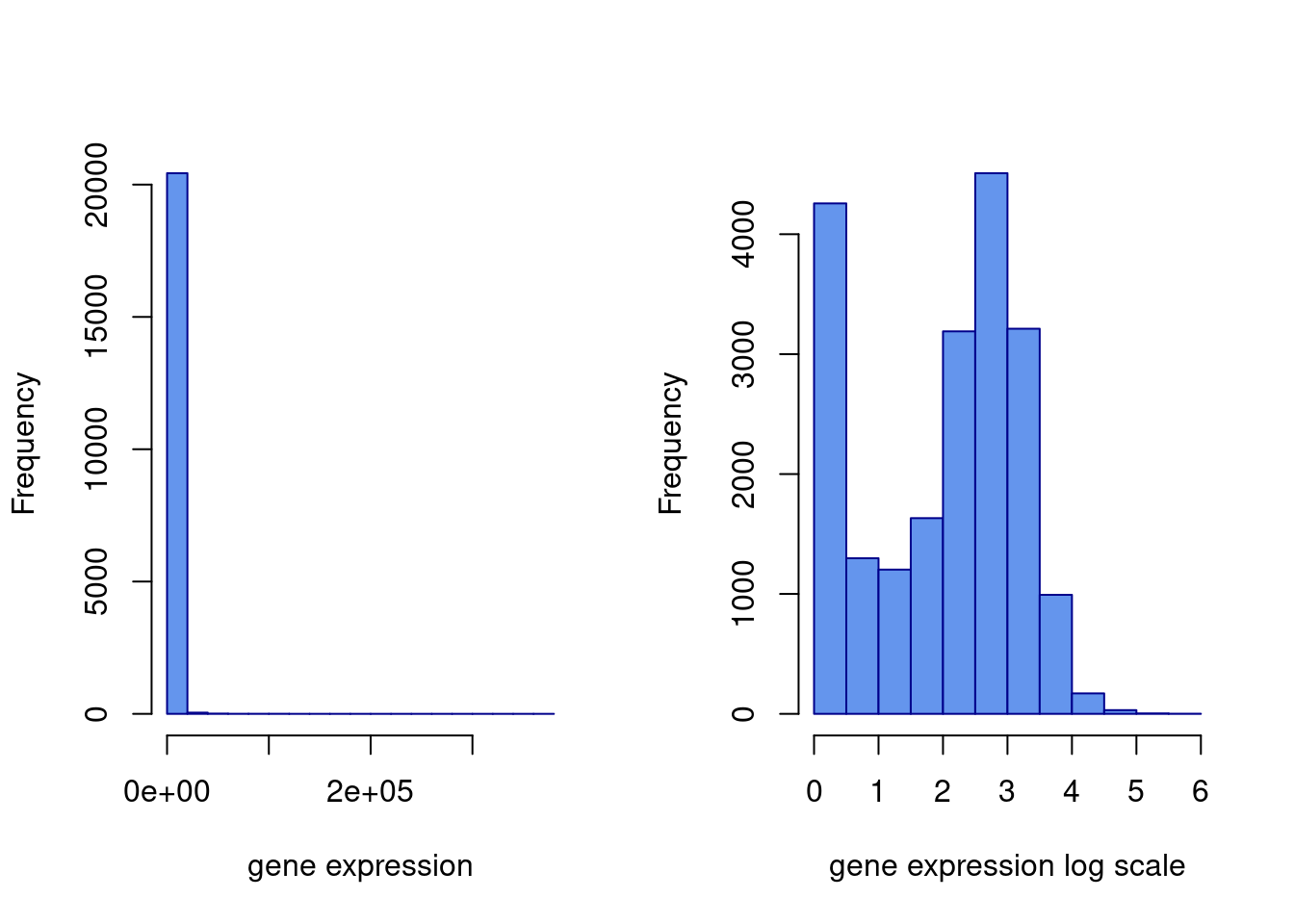
Transform –> filter/scale –> handle missing values
Remove near zero variation for the columns at least b/c likely not to have predictive value
library(caret)
# 85% of the values are the same
# this function creates the filter but doesn't apply it yet
nzv=preProcess(tgexp,method="nzv",uniqueCut = 15)
# apply the filter using "predict" function
# return the filtered dataset and assign it to nzv_tgexp variable
nzv_tgexp=predict(nzv,tgexp)How many variable predictors? This can be arbitrary
What about centering and scaling the data?
Filter out highly correlated variables so the model is fitted faster (optional depending on the type of analysis)
Transform –> filter/scale –> handle missing values
NA is NOT ZERO!!
Options for dealing with missing values … it depends. -Discard samples and/or predictors with missing values -Impute missing values via algorithm
#add an NA to the dataset
missing_tgexp=tgexp
missing_tgexp[1,1]=NA
#remove from data set
gexpnoNA=missing_tgexp[,colSums(is.na(missing_tgexp)) == 0]
#impute missing value w/ caret::preProcess
library(caret)
mImpute=preProcess(missing_tgexp,method="medianImpute")
imputedGexp=predict(mImpute,missing_tgexp)
#another, possibly more accurate imputation via nearest neighbors
library(RANN)
knnImpute=preProcess(missing_tgexp,method="knnImpute")
knnimputedGexp=predict(knnImpute,missing_tgexp)