5.10 Prediciting continuous variables by regression
# file path for CpG methylation and age
fileMethAge=system.file("extdata",
"CpGmeth2Age.rds",
package="compGenomRData")
# read methylation-age table
ameth=readRDS(fileMethAge)
dim(ameth)## [1] 108 27579## Age cg26211698 cg03790787
## Min. :-0.4986 Min. :0.01223 Min. :0.05001
## 1st Qu.:-0.4027 1st Qu.:0.01885 1st Qu.:0.07818
## Median :18.8466 Median :0.02269 Median :0.08964
## Mean :25.9083 Mean :0.02483 Mean :0.09300
## 3rd Qu.:49.6110 3rd Qu.:0.02888 3rd Qu.:0.10423
## Max. :83.6411 Max. :0.04883 Max. :0.16271# plot histogram of methylation values
hist(unlist(ameth[,-1]),border="white",
col="cornflowerblue",main="",xlab="methylation values")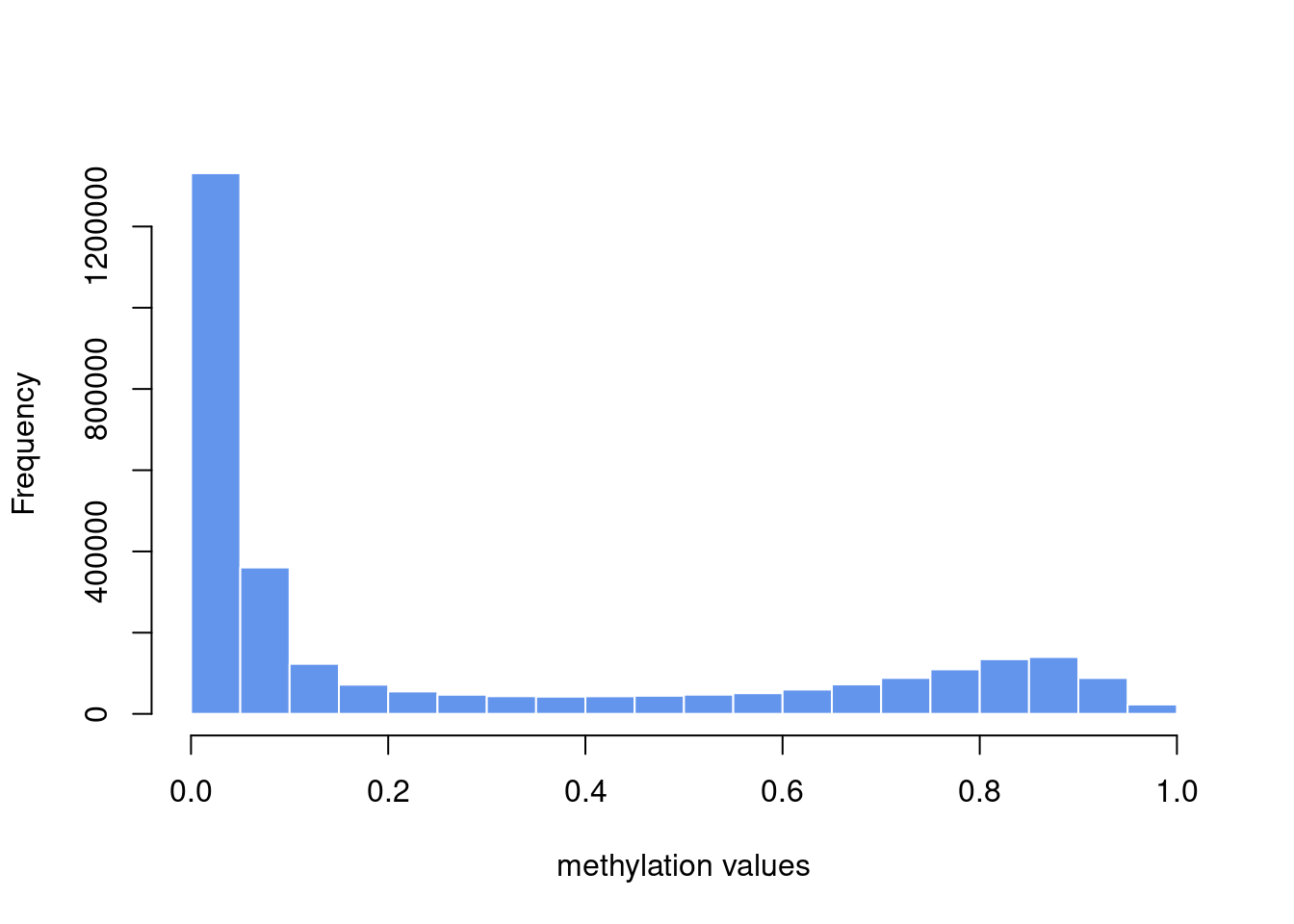
# remove CpGs with less than 0.1 StDev
ameth=ameth[,c(TRUE,matrixStats::colSds(as.matrix(ameth[,-1]))>0.1)]
dim(ameth)## [1] 108 2290Running random forest regression
set.seed(18)
par(mfrow=c(1,2))
# we are not going to do any cross-validation
# and rely on OOB error
trctrl <- trainControl(method = "none")
# we will now train random forest model
rfregFit <- train(Age~.,
data = ameth,
method = "ranger",
trControl=trctrl,
# calculate importance
importance="permutation",
tuneGrid = data.frame(mtry=50,
min.node.size = 5,
splitrule="variance")
)
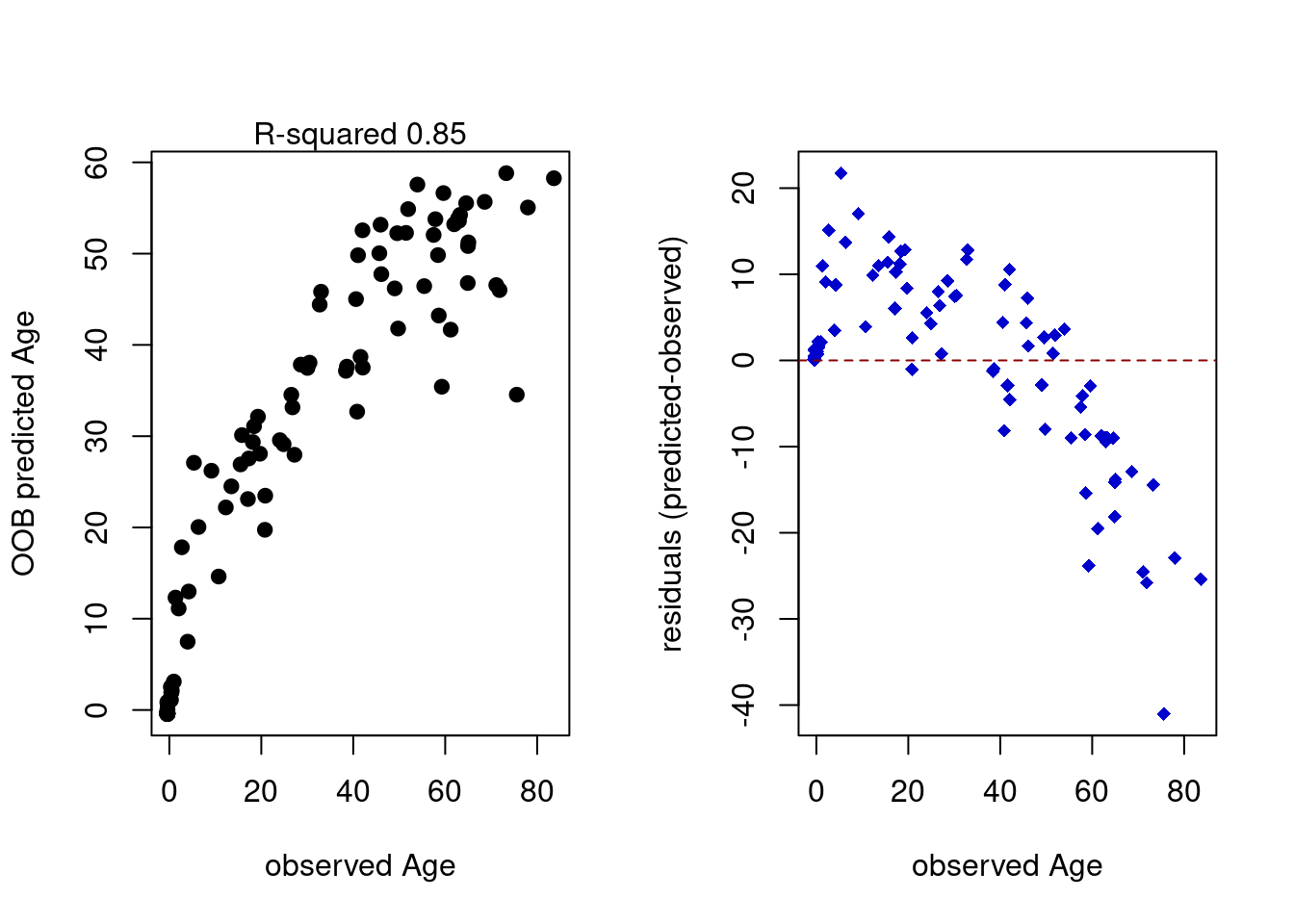
# plot Observed vs OOB predicted values from the model
plot(ameth$Age,rfregFit$finalModel$predictions,
pch=19,xlab="observed Age",
ylab="OOB predicted Age")
mtext(paste("R-squared",
format(rfregFit$finalModel$r.squared,digits=2)))
# plot residuals
plot(ameth$Age,(rfregFit$finalModel$predictions-ameth$Age),
pch=18,ylab="residuals (predicted-observed)",
xlab="observed Age",col="blue3")
abline(h=0,col="red4",lty=2)