5.9 Other algorithms
Gradient boosting
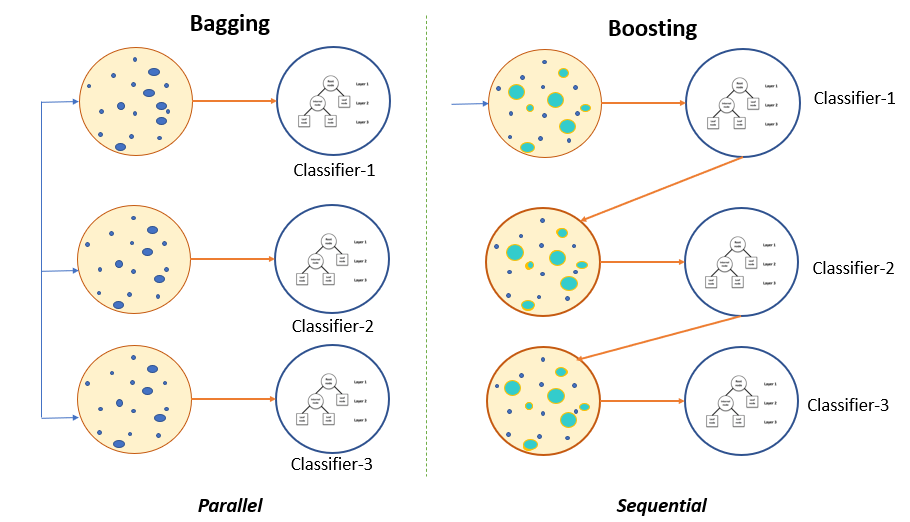
Gradient boosting. Source: http://haris.agaramsolutions.com/xwh/gradient-boosting-explained.html
##
## Attaching package: 'xgboost'## The following object is masked from 'package:dplyr':
##
## sliceset.seed(17)
# we will just set up 5-fold cross validation
trctrl <- trainControl(method = "cv",number=5)
# we will now train elastic net model
# it will try
gbFit <- train(subtype~., data = training,
method = "xgbTree",
trControl=trctrl,
# paramters to try
tuneGrid = data.frame(nrounds=200,
eta=c(0.05,0.1,0.3),
max_depth=4,
gamma=0,
colsample_bytree=1,
subsample=0.5,
min_child_weight=1))
# best parameters by cross-validation accuracy
gbFit## eXtreme Gradient Boosting
##
## 130 samples
## 804 predictors
## 2 classes: 'CIMP', 'noCIMP'
##
## No pre-processing
## Resampling: Cross-Validated (5 fold)
## Summary of sample sizes: 104, 104, 104, 104, 104
## Resampling results across tuning parameters:
##
## eta Accuracy Kappa
## 0.05 0.9846154 0.9692308
## 0.10 0.9846154 0.9692308
## 0.30 0.9692308 0.9384615
##
## Tuning parameter 'nrounds' was held constant at a value of 200
## Tuning
##
## Tuning parameter 'min_child_weight' was held constant at a value of 1
##
## Tuning parameter 'subsample' was held constant at a value of 0.5
## Accuracy was used to select the optimal model using the largest value.
## The final values used for the model were nrounds = 200, max_depth = 4, eta
## = 0.05, gamma = 0, colsample_bytree = 1, min_child_weight = 1 and subsample
## = 0.5.## nrounds max_depth eta gamma colsample_bytree min_child_weight subsample
## 1 200 4 0.05 0 1 1 0.5Support Vector Machines
##
## Attaching package: 'kernlab'## The following object is masked from 'package:mosaic':
##
## cross## The following object is masked from 'package:ggplot2':
##
## alphaset.seed(17)
# we will just set up 5-fold cross validation
trctrl <- trainControl(method = "cv",number=5)
# we will now train elastic net model
# it will try
svmFit <- train(subtype~., data = training,
# this SVM used radial basis function
method = "svmRadial",
trControl=trctrl,
tuneGrid=data.frame(C=c(0.25,0.5,1),
sigma=1))
svmFit## Support Vector Machines with Radial Basis Function Kernel
##
## 130 samples
## 804 predictors
## 2 classes: 'CIMP', 'noCIMP'
##
## No pre-processing
## Resampling: Cross-Validated (5 fold)
## Summary of sample sizes: 104, 104, 104, 104, 104
## Resampling results across tuning parameters:
##
## C Accuracy Kappa
## 0.25 0.6846154 0.3692308
## 0.50 0.6846154 0.3692308
## 1.00 0.6923077 0.3846154
##
## Tuning parameter 'sigma' was held constant at a value of 1
## Accuracy was used to select the optimal model using the largest value.
## The final values used for the model were sigma = 1 and C = 1.## sigma C
## 3 1 1Neural Networks
library(nnet)
set.seed(17)
# we will just set up 5-fold cross validation
trctrl <- trainControl(method = "cv",number=5)
# we will now train neural net model
# it will try
nnetFit <- train(subtype~., data = training,
method = "nnet",
trControl=trctrl,
tuneGrid=data.frame(size=1:2,decay=0
),
# this is maximum number of weights
# needed for the nnet method
MaxNWts=2000) ## # weights: 807
## initial value 75.647538
## final value 68.770195
## converged
## # weights: 1613
## initial value 65.383006
## final value 44.328983
## converged
## # weights: 807
## initial value 83.888393
## final value 71.777468
## converged
## # weights: 1613
## initial value 73.951723
## final value 59.258217
## converged
## # weights: 807
## initial value 76.998643
## final value 71.761891
## converged
## # weights: 1613
## initial value 81.545396
## final value 70.423633
## converged
## # weights: 807
## initial value 73.329419
## final value 71.767420
## converged
## # weights: 1613
## initial value 75.060274
## final value 66.896640
## converged
## # weights: 807
## initial value 77.715686
## final value 69.628207
## converged
## # weights: 1613
## initial value 73.978955
## final value 69.194435
## converged
## # weights: 1613
## initial value 86.959405
## final value 80.223758
## converged## Neural Network
##
## 130 samples
## 804 predictors
## 2 classes: 'CIMP', 'noCIMP'
##
## No pre-processing
## Resampling: Cross-Validated (5 fold)
## Summary of sample sizes: 104, 104, 104, 104, 104
## Resampling results across tuning parameters:
##
## size Accuracy Kappa
## 1 0.5923077 0.1846154
## 2 0.7076923 0.4153846
##
## Tuning parameter 'decay' was held constant at a value of 0
## Accuracy was used to select the optimal model using the largest value.
## The final values used for the model were size = 2 and decay = 0.## size decay
## 2 2 0Ensemble Models
# predict with k-NN model
knnPred=as.character(predict(knnFit,testing[,-1],type="class"))
# predict with elastic Net model
enetPred=as.character(predict(enetFit,testing[,-1]))
# predict with random forest model
rfPred=as.character(predict(rfFit,testing[,-1]))
# do voting for class labels
# code finds the most frequent class label per row
votingPred=apply(cbind(knnPred,enetPred,rfPred),1,
function(x) names(which.max(table(x))))
# check accuracy
confusionMatrix(data=testing[,1],
reference=as.factor(votingPred))$overall[1]## Accuracy
## 0.9074074