8.7 Accounting for additional sources of variation
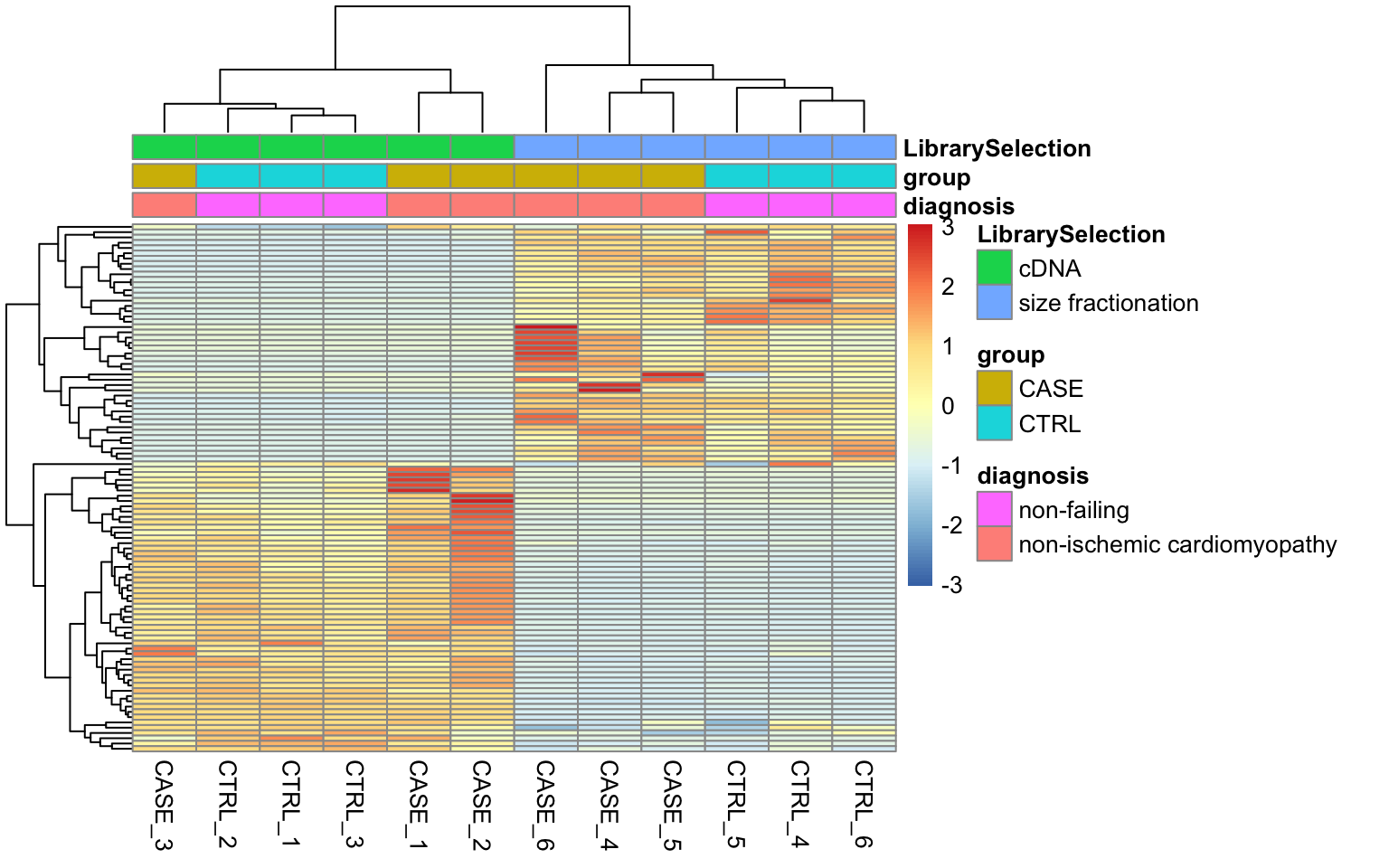
batcheffect
library(DESeq2)
# remove the 'width' column from the counts matrix
countData <- as.matrix(subset(counts, select = c(-width)))
# set up a DESeqDataSet object
dds <- DESeqDataSetFromMatrix(countData = countData,
colData = colData,
design = ~ LibrarySelection + group)IMPORTANT: keep the variable of interest at the last position in design formula.
Unknown source of variation –> RUVSeq or sva
RUVg() uses a set of reference genes, that shouldn’t change due to the condition.
RUVs() estimates the correction factor by assuming that replicates should have constant biological variation