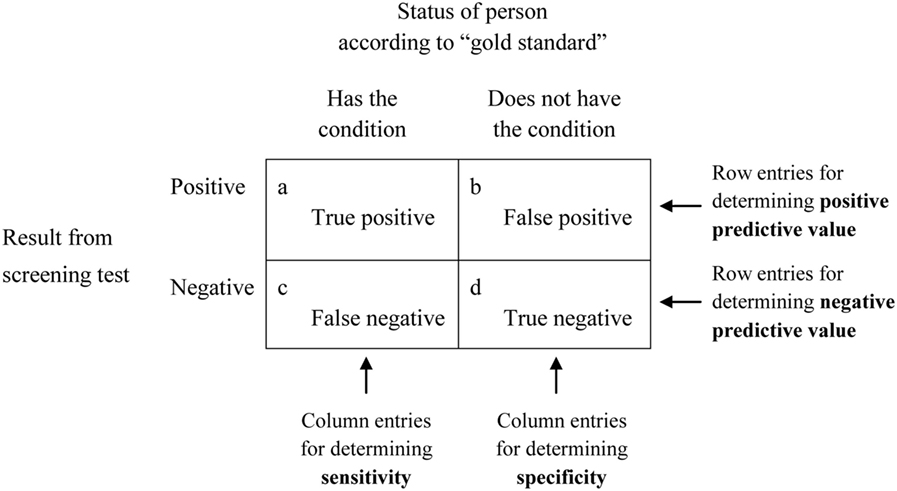
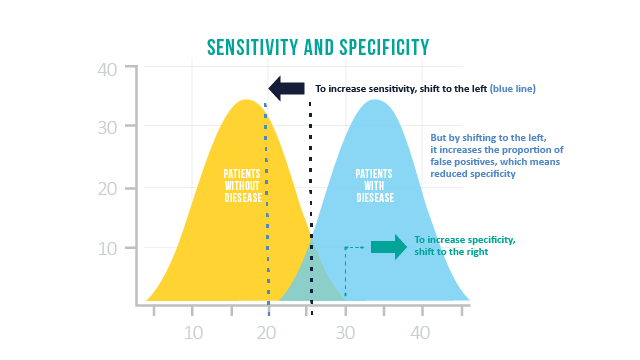
5.5 Precision, Specificity, and Sensitivity
# get k-NN prediction on the training data itself, with k=5
knnFit=knn3(x=training[,-1], # training set
y=training[,1], # training set class labels
k=5)
# predictions on the training set
trainPred=predict(knnFit,training[,-1],type="class")
# compare the predicted labels to real labels
# get different performance metrics
confusionMatrix(data=training[,1],reference=trainPred)## Confusion Matrix and Statistics
##
## Reference
## Prediction CIMP noCIMP
## CIMP 62 3
## noCIMP 13 52
##
## Accuracy : 0.8769
## 95% CI : (0.8078, 0.928)
## No Information Rate : 0.5769
## P-Value [Acc > NIR] : 9.216e-14
##
## Kappa : 0.7538
##
## Mcnemar's Test P-Value : 0.02445
##
## Sensitivity : 0.8267
## Specificity : 0.9455
## Pos Pred Value : 0.9538
## Neg Pred Value : 0.8000
## Prevalence : 0.5769
## Detection Rate : 0.4769
## Detection Prevalence : 0.5000
## Balanced Accuracy : 0.8861
##
## 'Positive' Class : CIMP
## # predictions on the test set, return class labels
testPred=predict(knnFit,testing[,-1],type="class")
# compare the predicted labels to real labels
# get different performance metrics
confusionMatrix(data=testing[,1],reference=testPred)## Confusion Matrix and Statistics
##
## Reference
## Prediction CIMP noCIMP
## CIMP 25 2
## noCIMP 6 21
##
## Accuracy : 0.8519
## 95% CI : (0.7288, 0.9338)
## No Information Rate : 0.5741
## P-Value [Acc > NIR] : 1.182e-05
##
## Kappa : 0.7037
##
## Mcnemar's Test P-Value : 0.2888
##
## Sensitivity : 0.8065
## Specificity : 0.9130
## Pos Pred Value : 0.9259
## Neg Pred Value : 0.7778
## Prevalence : 0.5741
## Detection Rate : 0.4630
## Detection Prevalence : 0.5000
## Balanced Accuracy : 0.8597
##
## 'Positive' Class : CIMP
## ## Type 'citation("pROC")' for a citation.##
## Attaching package: 'pROC'## The following objects are masked from 'package:mosaic':
##
## cov, var## The following objects are masked from 'package:stats':
##
## cov, smooth, var# get k-NN class probabilities
# prediction probabilities on the test set
testProbs=predict(knnFit,testing[,-1])
# get the roc curve
rocCurve <- pROC::roc(response = testing[,1],
predictor = testProbs[,1],
## This function assumes that the second
## class is the class of interest, so we
## reverse the labels.
levels = rev(levels(testing[,1])))## Setting direction: controls < cases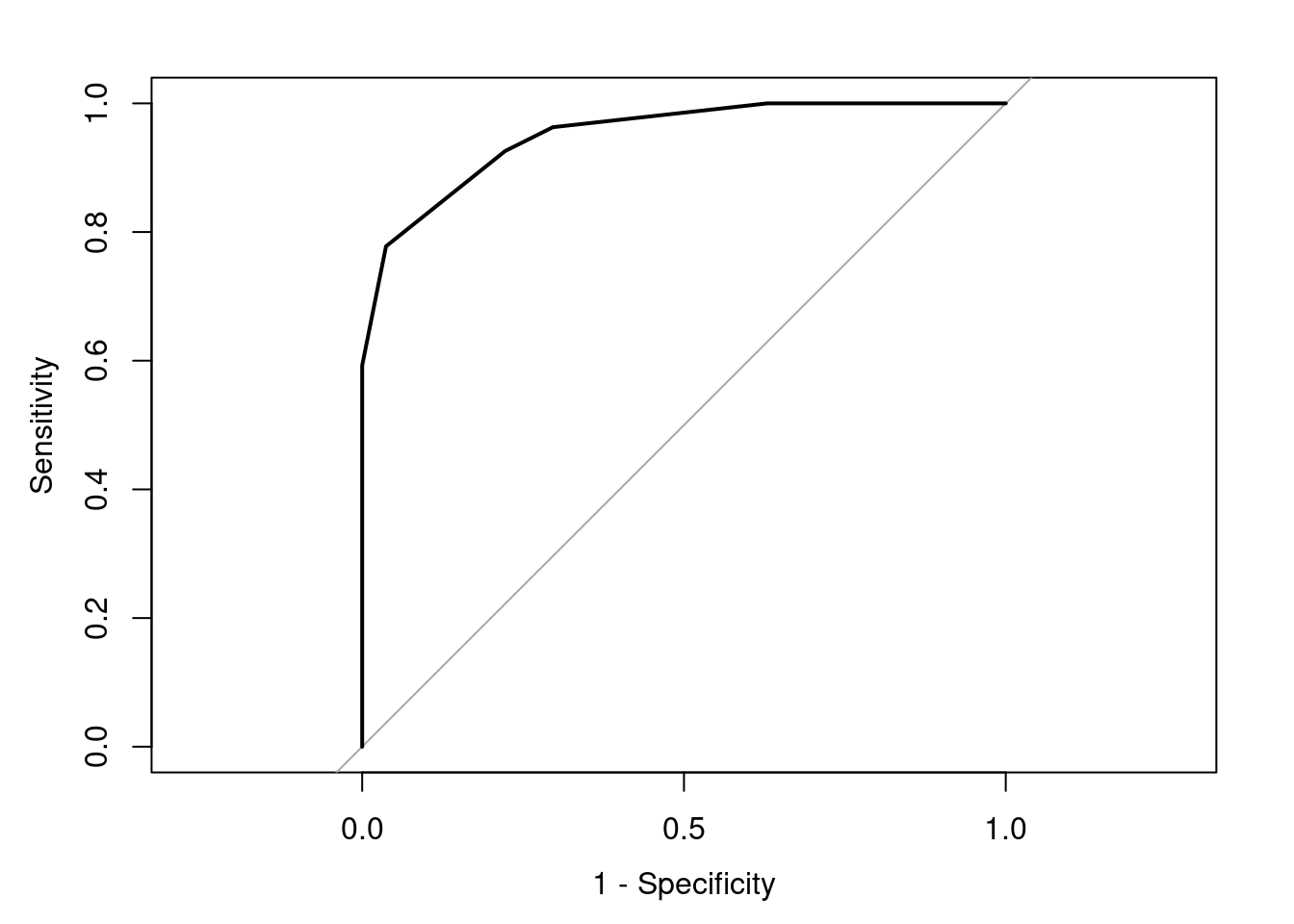
## Area under the curve: 0.9506