7.3 tidyverse
Package sf has tidyverse-style read and write functions, read_sf and write_sf that
- return a tibble rather than a data.frame
- do not print any output
- overwrite existing data by default
The dplyr, ggplot2, and tidyr capabilities are probably familiar for this audience, but there are some extra considerations for sf data below.
7.3.1 summarise
The summarise method for sf objects has two special arguments:
do_union(defaultTRUE) determines whether grouped geometries are unioned on return, so that they form a valid geometryis_coverage(defaultFALSE) in case the geometries grouped form a coverage (do not have overlaps), setting this toTRUEspeeds up the unioning
7.3.2 filter
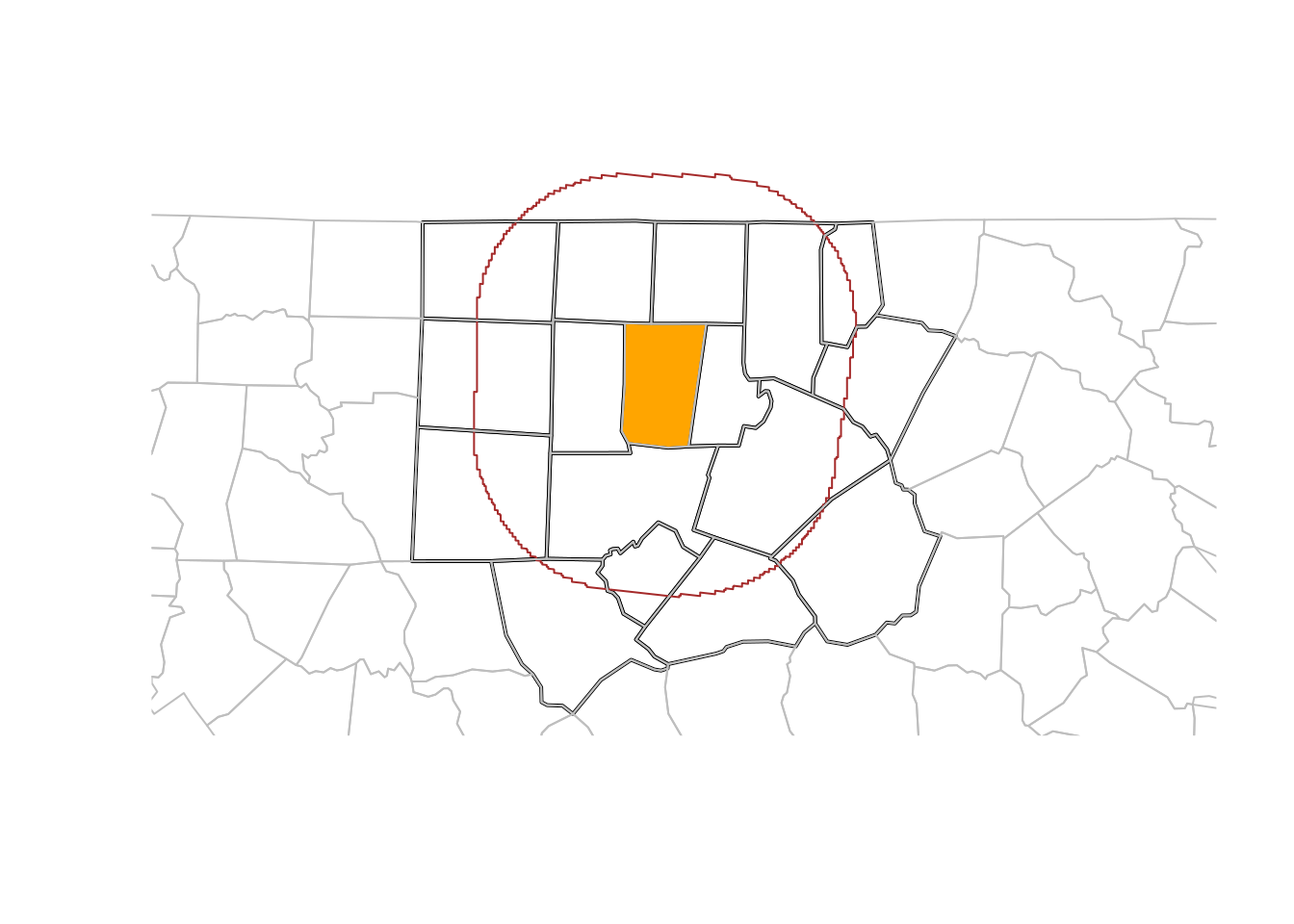
We can use filter for spatial predicates. For example, to select all counties less than 50 km away from Orange County,
# filters
orange <- nc |> dplyr::filter(NAME == "Orange")
wd <- st_is_within_distance(nc, orange,
units::set_units(50, km))
o50 <- nc |> dplyr::filter(lengths(wd) > 0)
# plot
og <- st_geometry(orange)
buf50 <- st_buffer(og, units::set_units(50, km))
all <- c(buf50, st_geometry(o50))
plot(st_geometry(o50), lwd = 2, extent = all)
plot(og, col = 'orange', add = TRUE)
plot(buf50, add = TRUE, col = NA, border = 'brown')
plot(st_geometry(nc), add = TRUE, border = 'grey')