15.1 Theory
Tobler’s first law of geography
“Everything is related to everything else, but near things are more related than distant things”
Recalling John Snow's belief that the cause of cholera was water-borne.
In summary, spatial autocorrelation is the degree to which the values of a variable in one location are correlated with the values of the same variable in neighboring locations.
15.1.1 Moran’s I
Moran's Test \(I\) helps us understand if this similarity (nearby locations are more similar to each other than to distant locations) is statistically significant or just due to chance.
It will provide a p-value, if the p-value is low (typically less than 0.05), there is significant spatial autocorrelation in the data, or a non-random pattern.
\[I=\frac{n\sum_{(2)}{w_{ij}z_iz_j}}{S_0\sum_{i=1}^n{z_i^2}}\]
\(z_i=x_i-\bar{x}\)
\(\bar{x}=\sum_{i=1}^n{x_i/n}\)
\(w_{ij}=\text{spatial weights}\)
\(S_0= \sum_{(2)}{w_{ij}}\)
15.1.2 Case Study: Polish election data
The Polish election data 2015, pol_pres15, shows results of elections by type of area.
## Simple feature collection with 6 features and 2 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: 235157.1 ymin: 366913.3 xmax: 281431.7 ymax: 413016.6
## Projected CRS: ETRS89 / Poland CS92
## types I_turnout geometry
## 1 Urban 0.4768987 MULTIPOLYGON (((261089.5 38...
## 2 Rural 0.4338417 MULTIPOLYGON (((254150 3837...
## 3 Rural 0.4280679 MULTIPOLYGON (((275346 3846...
## 4 Urban/rural 0.3739735 MULTIPOLYGON (((251769.8 37...
## 5 Rural 0.4066215 MULTIPOLYGON (((263423.9 40...
## 6 Rural 0.3853731 MULTIPOLYGON (((267030.7 38...library(tidyverse)
map1 <- ggplot(pol_pres15) +
geom_sf(aes(geometry=geometry,fill=types),
linewidth=0.1,
color="grey80",
alpha=0.5)+
scale_fill_viridis_d(option = "C",direction = 1)+
labs(title="The Polish election data 2015",
subtitle = "observed",
fill="Types")+
ggthemes::theme_map()+
theme(legend.position = c(-0.2,0))
map1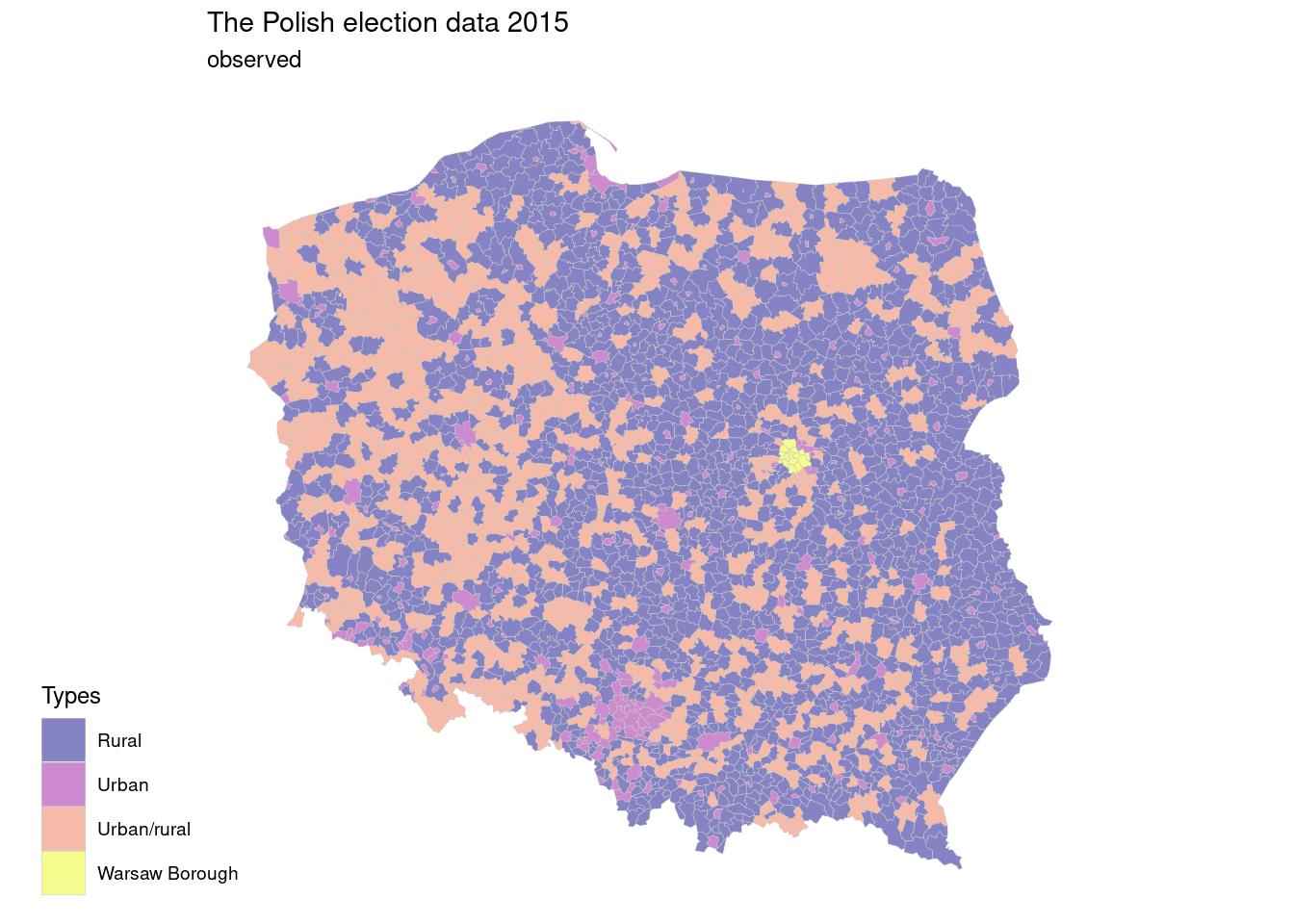
?st_make_valid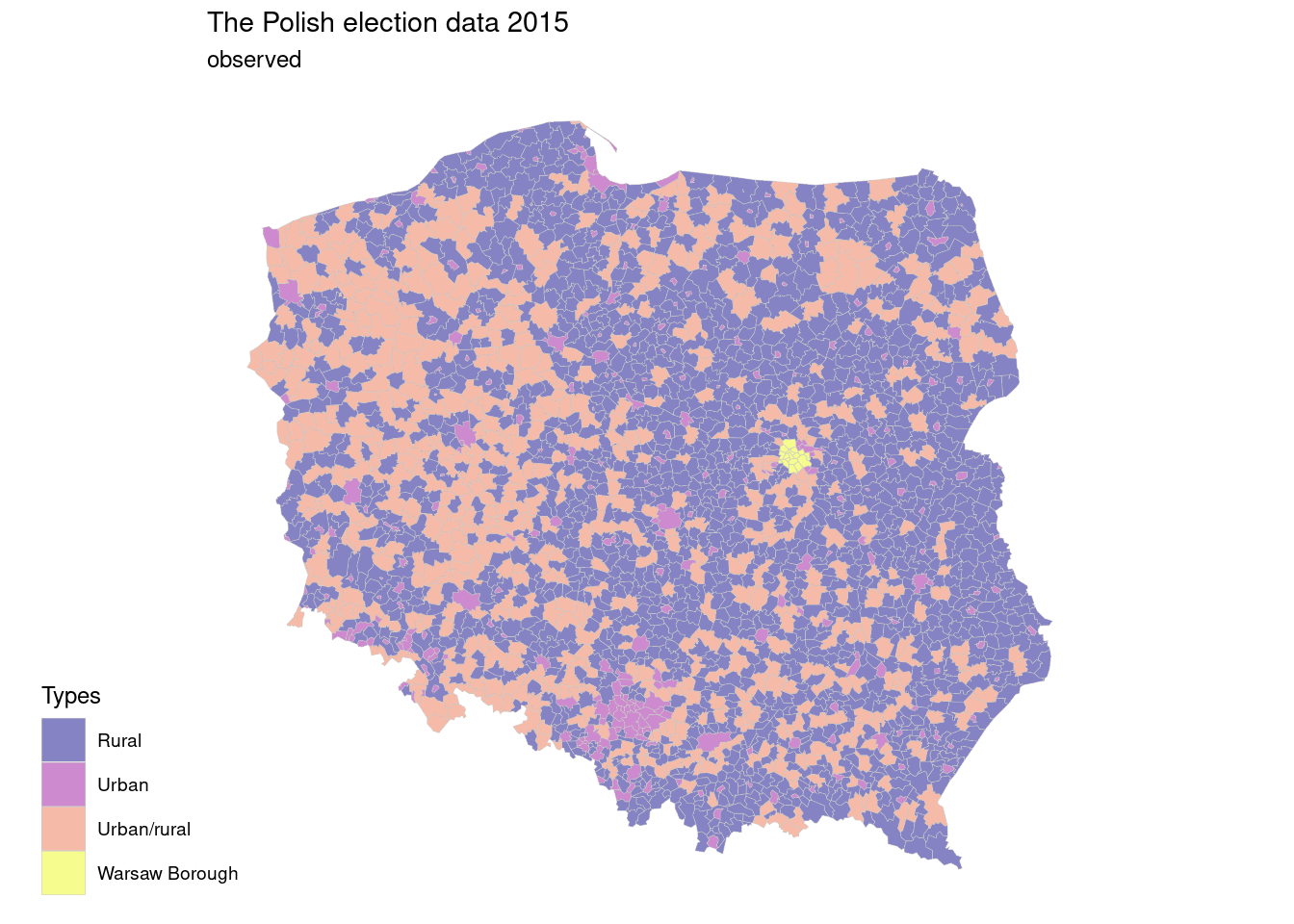
15.1.2.1 Moran’s I components
Construct neighbours list from polygon list based on regions with contiguous boundaries.
?poly2nb## Neighbour list object:
## Number of regions: 2495
## Number of nonzero links: 14242
## Percentage nonzero weights: 0.2287862
## Average number of links: 5.708216Spatial weights for neighbours lists function adds a weights list with values given by the coding scheme style chosen. B is the basic binary coding.
?nb2listw## Characteristics of weights list object:
## Neighbour list object:
## Number of regions: 2495
## Number of nonzero links: 14242
## Percentage nonzero weights: 0.2287862
## Average number of links: 5.708216
##
## Weights style: B
## Weights constants summary:
## n nn S0 S1 S2
## B 2495 6225025 14242 28484 35728015.1.2.2 Hypothesis testing
For testing for autocorrelation we first build a random Normal, rnorm distribution of values based on the number of observations.
Then apply the Moran’s I test for spatial autocorrelation to check the randomness of data.
?moran.test\[\left\{\begin{matrix} H_0 & p\leq0.05\\ H_a& p>0.05 \end{matrix}\right. \]
If the p-value is low (typically less than 0.05), it suggests that there is significant spatial autocorrelation in the data, indicating a non-random pattern.
##
## Moran I test under normality
##
## data: x
## weights: lw_q_B
##
## Moran I statistic standard deviate = -0.36932, p-value = 0.7119
## alternative hypothesis: two.sided
## sample estimates:
## Moran I statistic Expectation Variance
## -0.0047715202 -0.0004009623 0.0001400449Now see what happens if we use our data and add a gentle trend.
library(sf)
coords <- pol_pres15 |>
st_geometry() |> # select the geometry
st_centroid(of_largest_polygon = TRUE) Adding a gentle trend things changed:
beta <- 0.0015
t <- coords |>
st_coordinates() |> # transform the geometry into lat and long
subset(select = 1, drop = TRUE) |>
(function(x) x/1000)()This is our new data:
(x + beta * t) ##
## Moran I test under normality
##
## data: x_t
## weights: lw_q_B
##
## Moran I statistic standard deviate = 3.7015, p-value = 0.0002143
## alternative hypothesis: two.sided
## sample estimates:
## Moran I statistic Expectation Variance
## 0.0434026880 -0.0004009623 0.000140044915.1.3 Test the residuals
Here we apply the Moran’s I test for residual spatial autocorrelation to a linear model specification.
?lm.morantest##
## Global Moran I for regression residuals
##
## data:
## model: lm(formula = x_t ~ t)
## weights: lw_q_B
##
## Moran I statistic standard deviate = -0.33731, p-value = 0.7359
## alternative hypothesis: two.sided
## sample estimates:
## Observed Moran I Expectation Variance
## -0.0047772213 -0.0007891850 0.0001397877