10.7 Exercise 1
Use a random forest model to predict SID values (e.g. using package randomForest), and plot the random forest predictions against observations, along with the line.
## Simple feature collection with 6 features and 2 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -81.74107 ymin: 36.07282 xmax: -75.77316 ymax: 36.58965
## Geodetic CRS: NAD27
## # A tibble: 6 × 3
## SID rf geom
## <dbl> <dbl> <MULTIPOLYGON [°]>
## 1 0.000917 0.000666 (((-81.47276 36.23436, -81.54084 36.27251, -81.56198 36.273…
## 2 0 0.000518 (((-81.23989 36.36536, -81.24069 36.37942, -81.26284 36.405…
## 3 0.00157 0.00179 (((-80.45634 36.24256, -80.47639 36.25473, -80.53688 36.256…
## 4 0.00197 0.00110 (((-76.00897 36.3196, -76.01735 36.33773, -76.03288 36.3359…
## 5 0.00633 0.00442 (((-77.21767 36.24098, -77.23461 36.2146, -77.29861 36.2115…
## 6 0.00482 0.00479 (((-76.74506 36.23392, -76.98069 36.23024, -76.99475 36.235…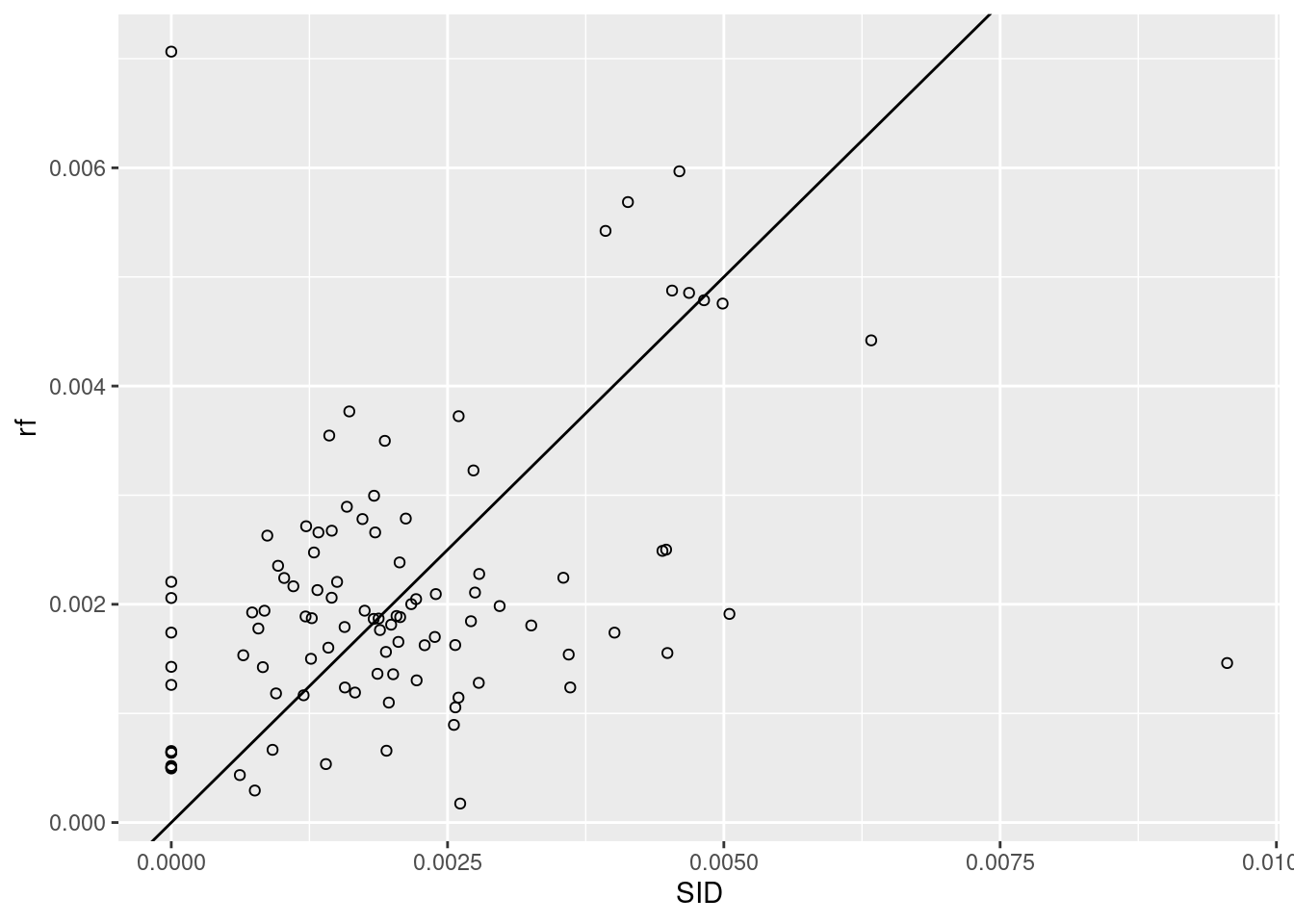
10.7.1 Exercise 2
Create a new dataset by randomly sampling 1000 points from the nc dataset, and rerun the linear regression model of section 10.2 on this dataset. What has changed?
## Warning: attribute variables are assumed to be spatially constant throughout
## all geometries##
## Call:
## lm(formula = SID ~ NWB, data = nc1)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.0033253 -0.0007411 -0.0000691 0.0005479 0.0062218
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.0006773 0.0002327 2.910 0.00447 **
## NWB 0.0043785 0.0006204 7.058 2.44e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.001288 on 98 degrees of freedom
## Multiple R-squared: 0.337, Adjusted R-squared: 0.3302
## F-statistic: 49.82 on 1 and 98 DF, p-value: 2.438e-10##
## Call:
## lm(formula = SID ~ NWB, data = nc3)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.0033493 -0.0007691 -0.0000141 0.0005032 0.0061979
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 7.140e-04 7.126e-05 10.02 <2e-16 ***
## NWB 4.358e-03 1.846e-04 23.60 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.001158 on 998 degrees of freedom
## Multiple R-squared: 0.3582, Adjusted R-squared: 0.3576
## F-statistic: 557 on 1 and 998 DF, p-value: < 2.2e-16the standard error has decreased with a factor 3 (sqrt(10))
lm(SID ~ NWB, nc1) |>
predict(nc1, interval = "prediction") -> pr1
lm(SID ~ NWB, nc3) |>
predict(nc1, interval = "prediction") -> pr2
mean(pr1[,"upr"] - pr1[,"lwr"])## [1] 0.005161177## [1] 0.004550376lm(SID ~ NWB, nc1) |>
predict(nc1, interval = "confidence") -> pr1
lm(SID ~ NWB, nc3) |>
predict(nc1, interval = "confidence") -> pr2
mean(pr1[,"upr"] - pr1[,"lwr"])## [1] 0.0007025904## [1] 0.0002021510.7.2 Exercise 3
Do the water-land classification using class::knn.
## stars object with 3 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## L7_ETMs.tif 1 54 69 68.91242 86 255
## dimension(s):
## from to offset delta refsys point x/y
## x 1 349 288776 28.5 SIRGAS 2000 / UTM zone 25S FALSE [x]
## y 1 352 9120761 -28.5 SIRGAS 2000 / UTM zone 25S FALSE [y]
## band 1 6 NA NA NA NA## xmin ymin xmax ymax
## 288776.3 9110728.8 298722.8 9120760.8## Geometry set for 1 feature
## Geometry type: POLYGON
## Dimension: XY
## Bounding box: xmin: 288776.3 ymin: 9110729 xmax: 298722.8 ymax: 9120761
## Projected CRS: SIRGAS 2000 / UTM zone 25S## POLYGON ((288776.3 9110729, 298722.8 9110729, 2...## stars object with 2 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## L7_ETMs.tif 12 41.75 63 60.95833 80.5 145
## dimension(s):
## from to refsys point
## geometry 1 20 SIRGAS 2000 / UTM zone 25S TRUE
## band 1 6 NA NA
## values
## geometry POINT (293002.2 9115516),...,POINT (290941.1 9114128)
## band NULL## [1] "stars"## x y band
## 349 352 6## Simple feature collection with 20 features and 6 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: 289053.6 ymin: 9110888 xmax: 298629.5 ymax: 9120451
## Projected CRS: SIRGAS 2000 / UTM zone 25S
## First 10 features:
## L7_ETMs.tif.V1 L7_ETMs.tif.V2 L7_ETMs.tif.V3 L7_ETMs.tif.V4 L7_ETMs.tif.V5
## 1 69 59 53 75 78
## 2 71 60 59 23 18
## 3 63 52 39 87 71
## 4 66 54 41 87 84
## 5 84 70 73 58 126
## 6 59 44 31 67 63
## 7 58 42 31 67 55
## 8 102 93 65 13 15
## 9 83 71 80 69 127
## 10 61 46 36 76 63
## L7_ETMs.tif.V6 geometry
## 1 45 POINT (293002.2 9115516)
## 2 13 POINT (290053.1 9110930)
## 3 33 POINT (292911.6 9119069)
## 4 42 POINT (292503.2 9116861)
## 5 102 POINT (291435.7 9113502)
## 6 30 POINT (294899.2 9116963)
## 7 27 POINT (291231.8 9118336)
## 8 13 POINT (298629.5 9112151)
## 9 110 POINT (295431.9 9119798)
## 10 33 POINT (292282 9118130)e_df <- st_as_sf(e) |>
st_coordinates() %>%
as_tibble() %>%
mutate(label=c(1:20),
group=ifelse(label%in%c(8, 14, 15, 18, 19),"land","water"))
e_df%>%head## # A tibble: 6 × 4
## X Y label group
## <dbl> <dbl> <int> <chr>
## 1 293002. 9115516. 1 water
## 2 290053. 9110930. 2 water
## 3 292912. 9119069. 3 water
## 4 292503. 9116861. 4 water
## 5 291436. 9113502. 5 water
## 6 294899. 9116963. 6 waterggplot()+
stars::geom_stars(data=r)+
geom_text(data=e_df,
mapping=aes(x=X,y=Y,
label=label,
group=group,
color=group),
check_overlap = T,
inherit.aes = F)+
scale_color_manual(values=c("red","yellow"))+
coord_equal()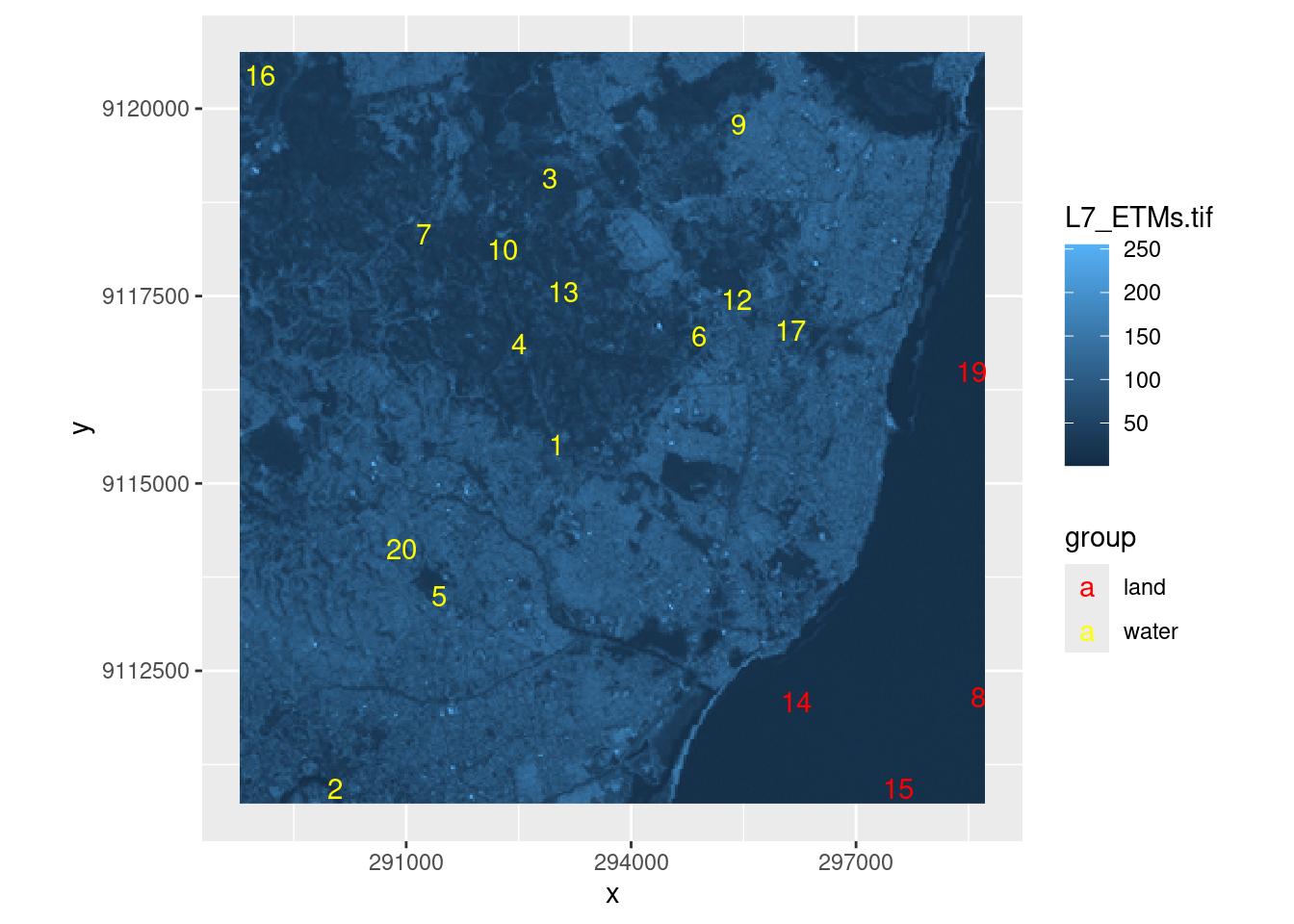
## stars object with 2 dimensions and 6 attributes
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## X1 47 67 78 79.14772 89 255
## X2 32 55 66 67.57465 79 255
## X3 21 49 63 64.35886 77 255
## X4 9 52 63 59.23541 75 255
## X5 1 63 89 83.18266 112 255
## X6 1 32 60 59.97521 88 255
## dimension(s):
## from to offset delta refsys point x/y
## x 1 349 288776 28.5 SIRGAS 2000 / UTM zone 25S FALSE [x]
## y 1 352 9120761 -28.5 SIRGAS 2000 / UTM zone 25S FALSE [y]library(class)
tr <- as.data.frame(trn) |> dplyr::select(X1, X2, X3, X4, X5, X6)
test <- as.data.frame(rs) |> dplyr::select(X1, X2, X3, X4, X5, X6) 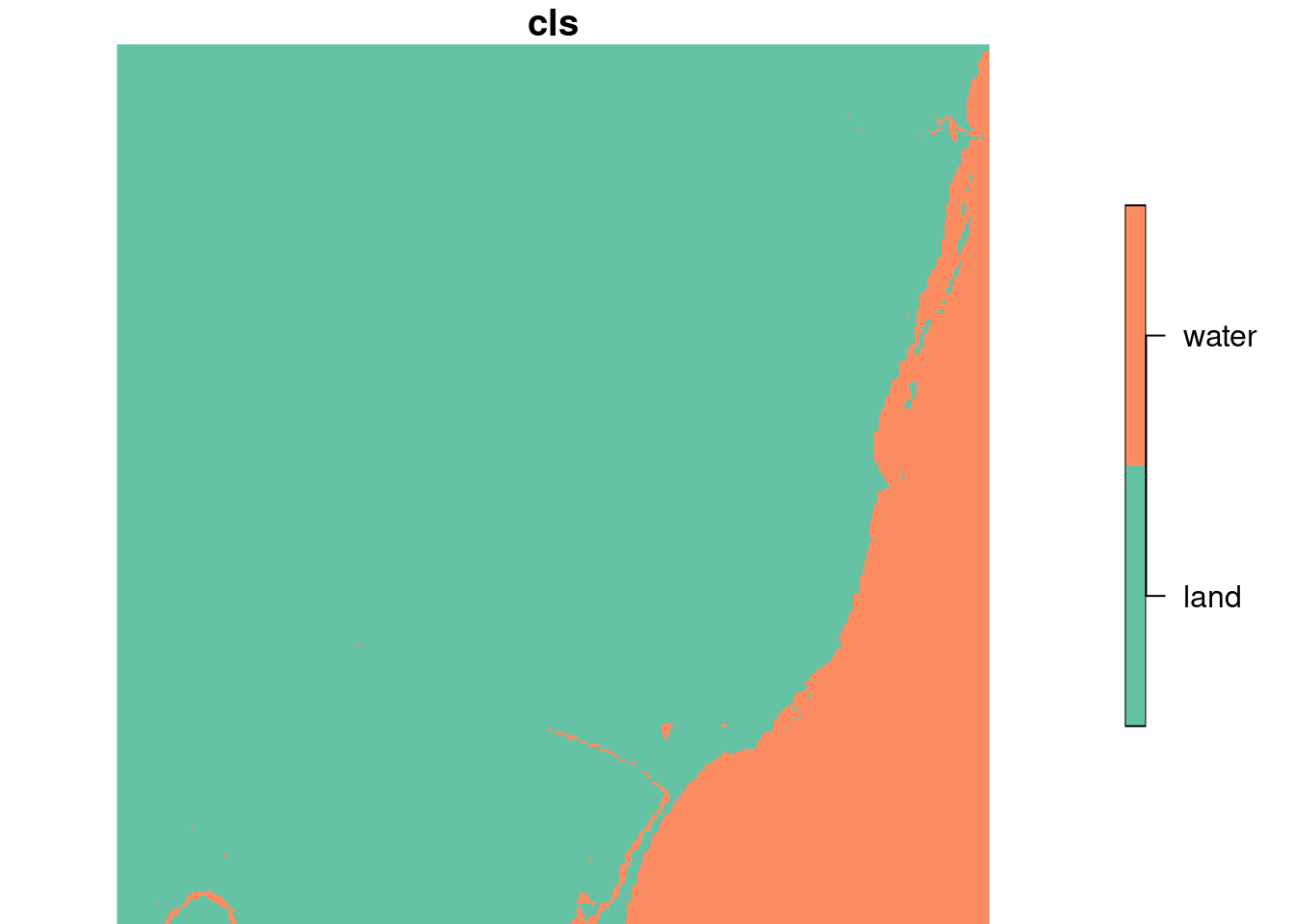
10.7.3 Exercise 4
For the linear model using nc and for the knn example of the previous exercise, add a first and a second order linear model in the spatial coordinates and compare the results (use st_centroid to obtain polygon centroids, and st_coordinates to extract the x and y coordinates in matrix form).
## Simple feature collection with 100 features and 17 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
## Geodetic CRS: NAD27
## # A tibble: 100 × 18
## AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74
## * <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <dbl> <dbl>
## 1 0.114 1.44 1825 1825 Ashe 37009 37009 5 1091 1 10
## 2 0.061 1.23 1827 1827 Alle… 37005 37005 3 487 0 10
## 3 0.143 1.63 1828 1828 Surry 37171 37171 86 3188 5 208
## 4 0.07 2.97 1831 1831 Curr… 37053 37053 27 508 1 123
## 5 0.153 2.21 1832 1832 Nort… 37131 37131 66 1421 9 1066
## 6 0.097 1.67 1833 1833 Hert… 37091 37091 46 1452 7 954
## 7 0.062 1.55 1834 1834 Camd… 37029 37029 15 286 0 115
## 8 0.091 1.28 1835 1835 Gates 37073 37073 37 420 0 254
## 9 0.118 1.42 1836 1836 Warr… 37185 37185 93 968 4 748
## 10 0.124 1.43 1837 1837 Stok… 37169 37169 85 1612 1 160
## # ℹ 90 more rows
## # ℹ 7 more variables: BIR79 <dbl>, SID79 <dbl>, NWBIR79 <dbl>,
## # geom <MULTIPOLYGON [°]>, SID <dbl>, NWB <dbl>, rf <dbl>## Warning: st_centroid assumes attributes are constant over geometries## X Y
## [1,] -81.49823 36.43140
## [2,] -81.12513 36.49111
## [3,] -80.68573 36.41252
## [4,] -76.02719 36.40714
## [5,] -77.41046 36.42236
## [6,] -76.99472 36.36142
## [7,] -76.23402 36.40122
## [8,] -76.70446 36.44428
## [9,] -78.11042 36.39693
## [10,] -80.23429 36.40042
## [11,] -79.33478 36.39346
## [12,] -79.77039 36.39610
## [13,] -78.65648 36.29993
## [14,] -78.97685 36.38870
## [15,] -78.41129 36.36224
## [16,] -77.65599 36.25302
## [17,] -76.31443 36.31231
## [18,] -81.15971 36.20158
## [19,] -81.69125 36.22481
## [20,] -76.45458 36.20486
## [21,] -76.61642 36.14886
## [22,] -81.92145 36.07194
## [23,] -80.66413 36.15481
## [24,] -78.28707 36.07760
## [25,] -80.25335 36.12571
## [26,] -79.78279 36.07305
## [27,] -79.39793 36.03757
## [28,] -76.98470 36.06189
## [29,] -79.12178 36.05355
## [30,] -78.87809 36.02862
## [31,] -77.98691 35.96228
## [32,] -82.16321 36.00591
## [33,] -77.59821 35.90717
## [34,] -81.54356 35.94703
## [35,] -82.30763 35.89215
## [36,] -77.10745 35.83637
## [37,] -78.65299 35.78443
## [38,] -82.70541 35.85323
## [39,] -80.87108 35.80259
## [40,] -80.54155 35.92495
## [41,] -81.17403 35.91575
## [42,] -80.20823 35.78986
## [43,] -81.70198 35.74484
## [44,] -76.58303 35.82508
## [45,] -76.21986 35.80954
## [46,] -82.04495 35.67528
## [47,] -79.80374 35.70551
## [48,] -79.25672 35.69645
## [49,] -77.92280 35.69918
## [50,] -80.51921 35.63691
## [51,] -77.37784 35.58906
## [52,] -81.21380 35.65816
## [53,] -82.52831 35.60684
## [54,] -78.36673 35.51338
## [55,] -82.98086 35.55136
## [56,] -75.80950 35.73533
## [57,] -76.87093 35.49589
## [58,] -83.48992 35.48069
## [59,] -77.67887 35.48228
## [60,] -79.17379 35.47106
## [61,] -81.91783 35.39871
## [62,] -78.00728 35.35727
## [63,] -78.87102 35.36660
## [64,] -81.54942 35.32956
## [65,] -81.22074 35.48086
## [66,] -83.13929 35.28244
## [67,] -79.47828 35.30615
## [68,] -80.82937 35.24481
## [69,] -80.55085 35.38494
## [70,] -79.90276 35.32716
## [71,] -80.24928 35.31388
## [72,] -82.47627 35.33467
## [73,] -83.82962 35.34507
## [74,] -77.64556 35.23278
## [75,] -82.79526 35.19968
## [76,] -81.17520 35.29245
## [77,] -82.17027 35.27570
## [78,] -83.42226 35.14516
## [79,] -78.36970 34.98647
## [80,] -76.76886 35.13564
## [81,] -84.05986 35.13111
## [82,] -78.82647 35.04375
## [83,] -77.35617 35.01856
## [84,] -80.53145 34.98630
## [85,] -80.10405 34.97520
## [86,] -79.23592 35.01163
## [87,] -76.24694 35.53213
## [88,] -77.93356 34.93235
## [89,] -79.74228 35.00140
## [90,] -83.74725 35.05384
## [91,] -77.10388 35.13065
## [92,] -79.47719 34.83945
## [93,] -77.44295 34.73908
## [94,] -79.10106 34.63869
## [95,] -76.69587 34.81997
## [96,] -78.55994 34.60928
## [97,] -77.91626 34.52688
## [98,] -78.65478 34.26335
## [99,] -77.89580 34.26162
## [100,] -78.25090 34.07671##
## Call:
## lm(formula = SID ~ NWB, data = nc1)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.0033253 -0.0007411 -0.0000691 0.0005479 0.0062218
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.0006773 0.0002327 2.910 0.00447 **
## NWB 0.0043785 0.0006204 7.058 2.44e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.001288 on 98 degrees of freedom
## Multiple R-squared: 0.337, Adjusted R-squared: 0.3302
## F-statistic: 49.82 on 1 and 98 DF, p-value: 2.438e-10##
## Call:
## lm(formula = SID ~ NWB + X + Y, data = nc4)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.0027669 -0.0007998 -0.0001568 0.0006015 0.0053235
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -1.248e-03 1.050e-02 -0.119 0.90564
## NWB 5.950e-03 7.983e-04 7.454 3.99e-11 ***
## X -2.266e-04 7.794e-05 -2.907 0.00453 **
## Y -4.655e-04 2.167e-04 -2.148 0.03424 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.001213 on 96 degrees of freedom
## Multiple R-squared: 0.424, Adjusted R-squared: 0.406
## F-statistic: 23.56 on 3 and 96 DF, p-value: 1.645e-11##
## Call:
## lm(formula = SID ~ NWB + X + Y + I(X^2) + I(Y^2) + X * Y, data = nc4)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.0027759 -0.0007526 -0.0001441 0.0006399 0.0053102
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 4.703e-01 7.119e-01 0.661 0.511
## NWB 5.978e-03 9.463e-04 6.317 9.08e-09 ***
## X -2.632e-04 1.016e-02 -0.026 0.979
## Y -2.719e-02 2.576e-02 -1.055 0.294
## I(X^2) -1.064e-05 3.387e-05 -0.314 0.754
## I(Y^2) 3.240e-04 3.445e-04 0.941 0.349
## X:Y -4.714e-05 1.688e-04 -0.279 0.781
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.001221 on 93 degrees of freedom
## Multiple R-squared: 0.4339, Adjusted R-squared: 0.3974
## F-statistic: 11.88 on 6 and 93 DF, p-value: 7.383e-10nc1$prediction_0 <- lm0 |> predict(nc4)
nc1$prediction_1 <- lm1 |> predict(nc4)
nc1$prediction_2 <- lm2 |> predict(nc4)nc5 <- nc1 %>%
dplyr::select(prediction_0,prediction_1,prediction_2) |>
#pivot_longer(cols = c("pr0","pr1","pr2"))%>%
st_as_stars() |>
merge()
nc5 ## stars object with 2 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median
## prediction_0.prediction_1.p... -0.0005071205 0.00127737 0.002034891
## Mean 3rd Qu. Max.
## prediction_0.prediction_1.p... 0.002045596 0.002776815 0.00517751
## dimension(s):
## from to refsys point
## geom 1 100 NAD27 FALSE
## attributes 1 3 NA NA
## values
## geom MULTIPOLYGON (((-81.47276...,...,MULTIPOLYGON (((-78.65572...
## attributes prediction_0, prediction_1, prediction_2ggplot()+
geom_stars(data=nc5)+
facet_wrap(~attributes,ncol = 1) +
scale_fill_viridis_c()+
labs(fill="")+
theme_void()+
theme(legend.position = "bottom",
legend.text = element_text(size=3))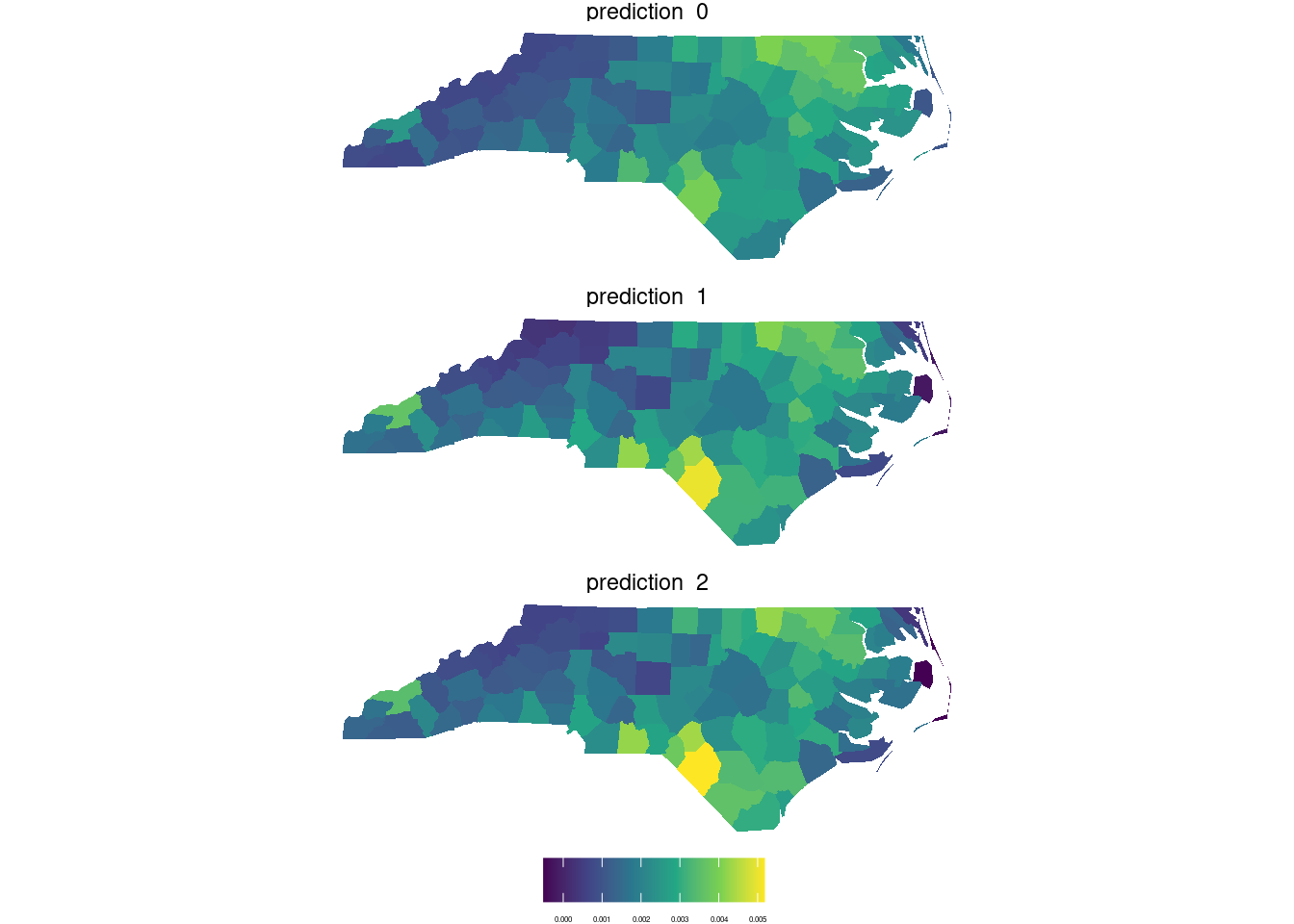
tr1 <- cbind(as.data.frame(trn),
st_coordinates(trn)) |>
dplyr::select(X, Y, X1, X2, X3, X4, X5, X6) ## stars object with 2 dimensions and 8 attributes
## attribute(s):
## X1 X2 X3 X4
## Min. : 47.00 Min. : 32.00 Min. : 21.00 Min. : 9.00
## 1st Qu.: 67.00 1st Qu.: 55.00 1st Qu.: 49.00 1st Qu.: 52.00
## Median : 78.00 Median : 66.00 Median : 63.00 Median : 63.00
## Mean : 79.15 Mean : 67.57 Mean : 64.36 Mean : 59.24
## 3rd Qu.: 89.00 3rd Qu.: 79.00 3rd Qu.: 77.00 3rd Qu.: 75.00
## Max. :255.00 Max. :255.00 Max. :255.00 Max. :255.00
## X5 X6 cls cls1
## Min. : 1.00 Min. : 1.00 land :102726 land :102318
## 1st Qu.: 63.00 1st Qu.: 32.00 water: 20122 water: 20530
## Median : 89.00 Median : 60.00
## Mean : 83.18 Mean : 59.98
## 3rd Qu.:112.00 3rd Qu.: 88.00
## Max. :255.00 Max. :255.00
## dimension(s):
## from to offset delta refsys point x/y
## x 1 349 288776 28.5 SIRGAS 2000 / UTM zone 25S FALSE [x]
## y 1 352 9120761 -28.5 SIRGAS 2000 / UTM zone 25S FALSE [y]tr2 <- cbind(as.data.frame(trn), st_coordinates(trn)) |>
transmute(X, Y, X2=X^2, Y2=Y^2, XY=X*Y, X1, X2, X3, X4, X5, X6)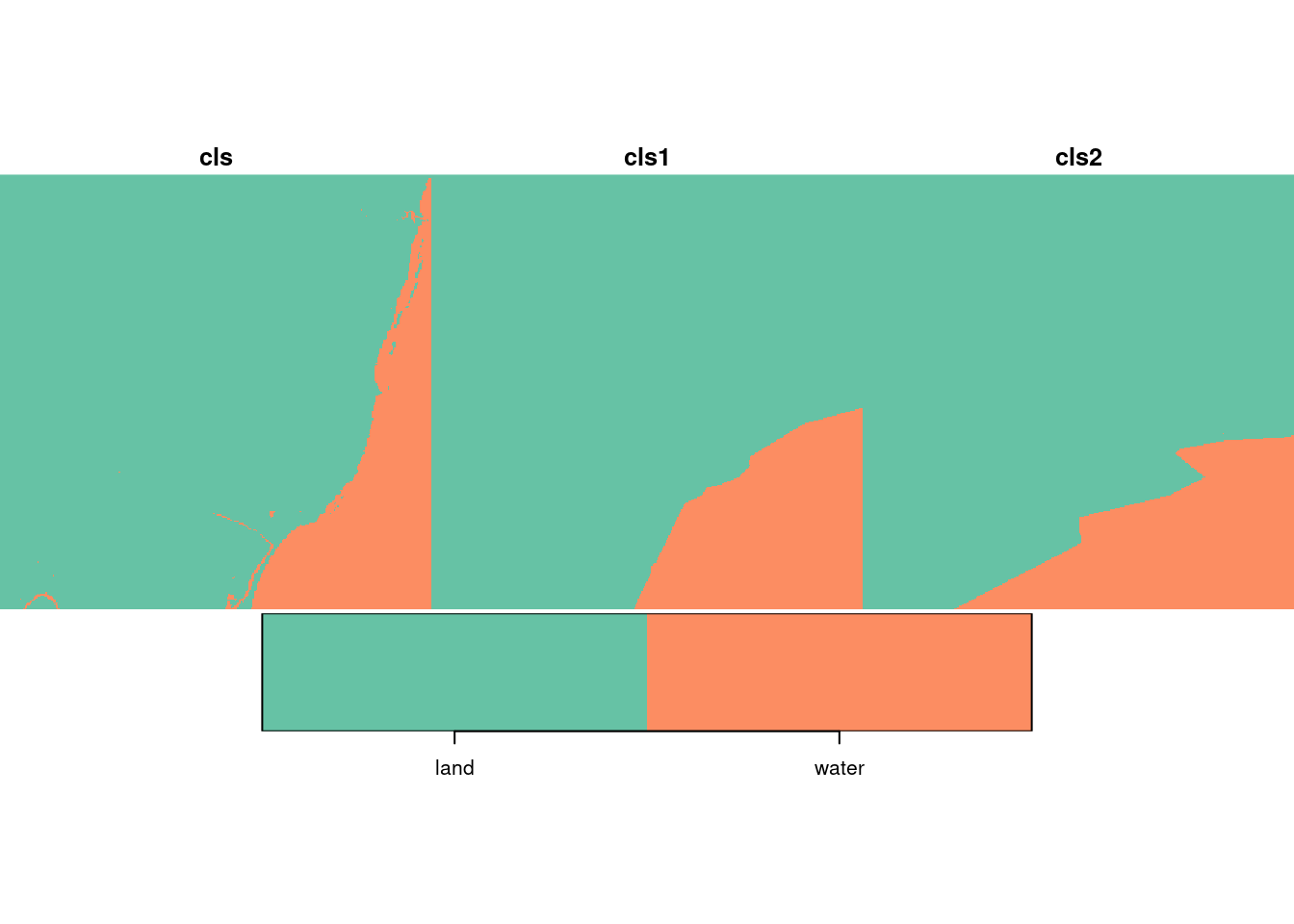
Soultions: https://edzer.github.io/sdsr_exercises/