Using fake data simulation to understand residual plots
Why do we plot residuals vs fitted values rather then observed values?
## # A tibble: 52 × 9
## hw1 hw2 hw3 hw4 midterm hw5 hw6 hw7 final
## <int> <int> <int> <int> <int> <int> <int> <int> <int>
## 1 95 88 100 95 80 96 99 0 103
## 2 0 74 74 0 53 83 97 0 79
## 3 100 0 105 100 91 96 100 96 122
## 4 0 90 76 100 63 91 95 0 78
## 5 100 96 99 100 91 93 100 92 135
## 6 90 83 95 100 73 89 100 90 117
## 7 95 98 100 100 59 98 98 94 135
## 8 80 100 97 100 69 94 98 101 123
## 9 95 90 98 90 78 95 99 100 109
## 10 90 94 95 98 91 94 100 89 126
## # ℹ 42 more rows## stan_glm
## family: gaussian [identity]
## formula: final ~ midterm
## observations: 52
## predictors: 2
## ------
## Median MAD_SD
## (Intercept) 64.2 17.7
## midterm 0.7 0.2
##
## Auxiliary parameter(s):
## Median MAD_SD
## sigma 14.9 1.6
##
## ------
## * For help interpreting the printed output see ?print.stanreg
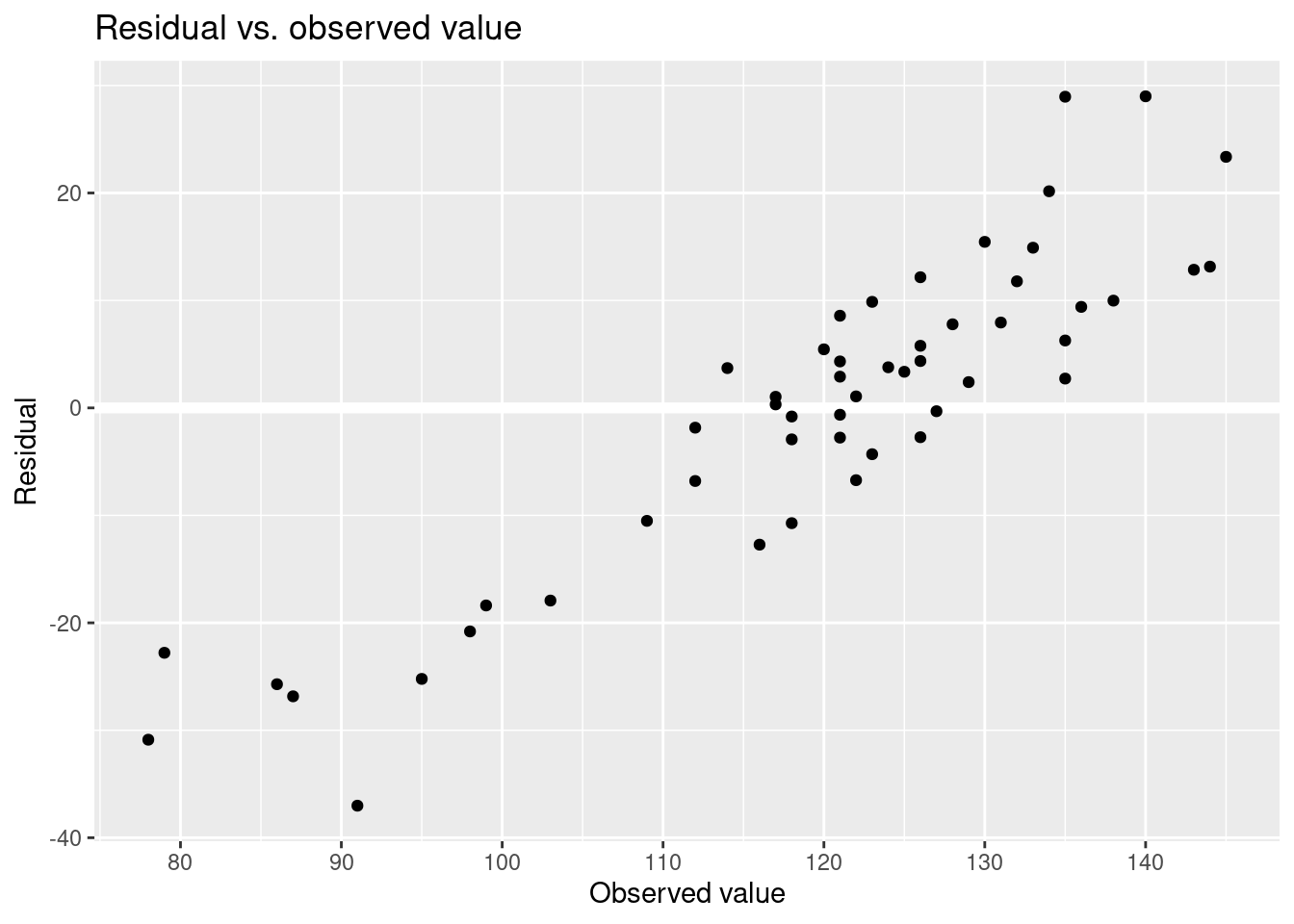
## * For info on the priors used see ?prior_summary.stanregPredicted values and residuals.
Residual vs. observed value.
Residual vs. predicted value.
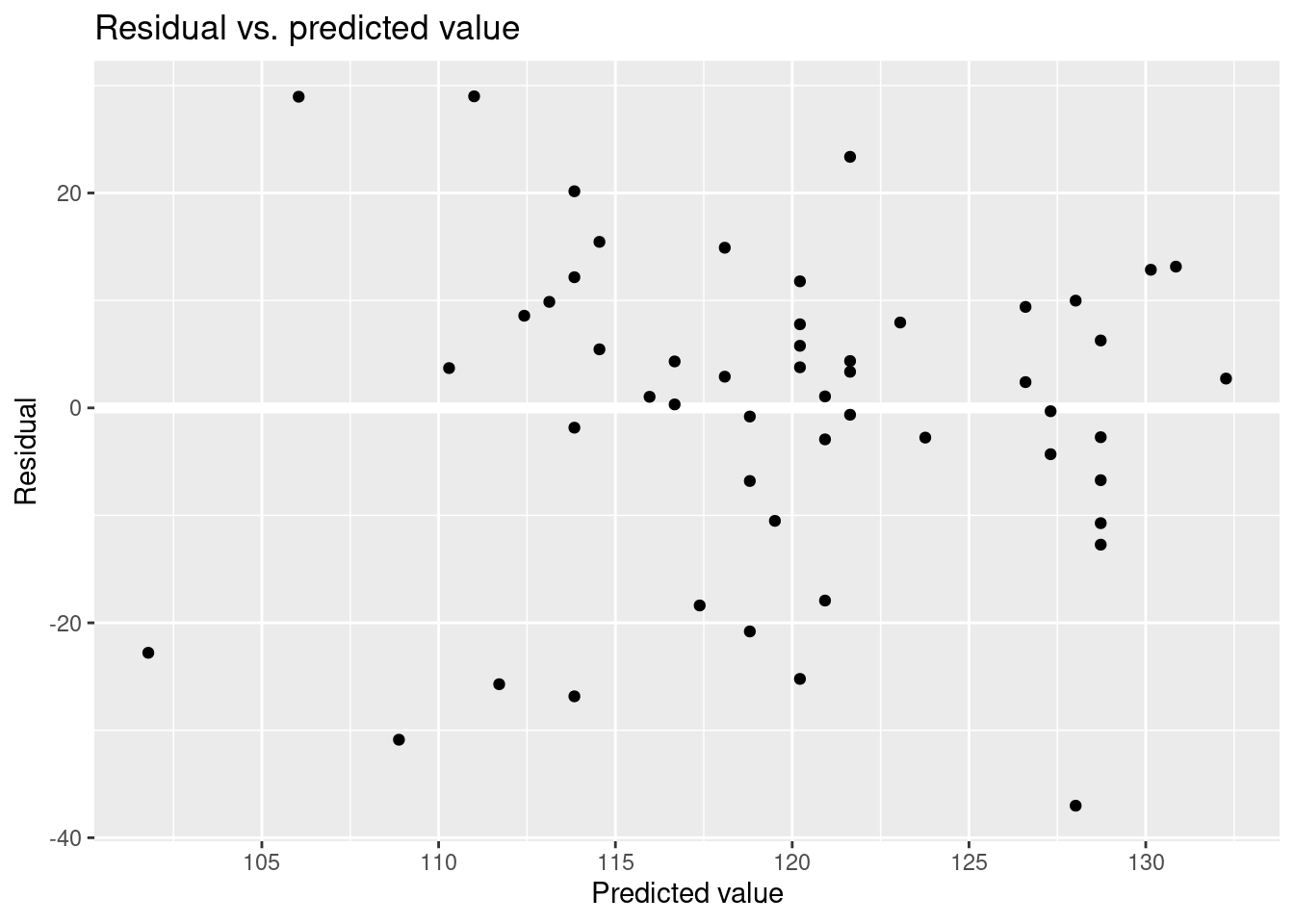
Understanding the choice using fake-data
set.seed(746)
intercept <- coef(fit)[["(Intercept)"]]
slope <- coef(fit)[["midterm"]]
sigma <- sigma(fit)
scores_sim <-
scores %>%
mutate(
pred = intercept + slope * midterm,
final_sim = pred + rnorm(n(), mean = 0, sd = sigma),
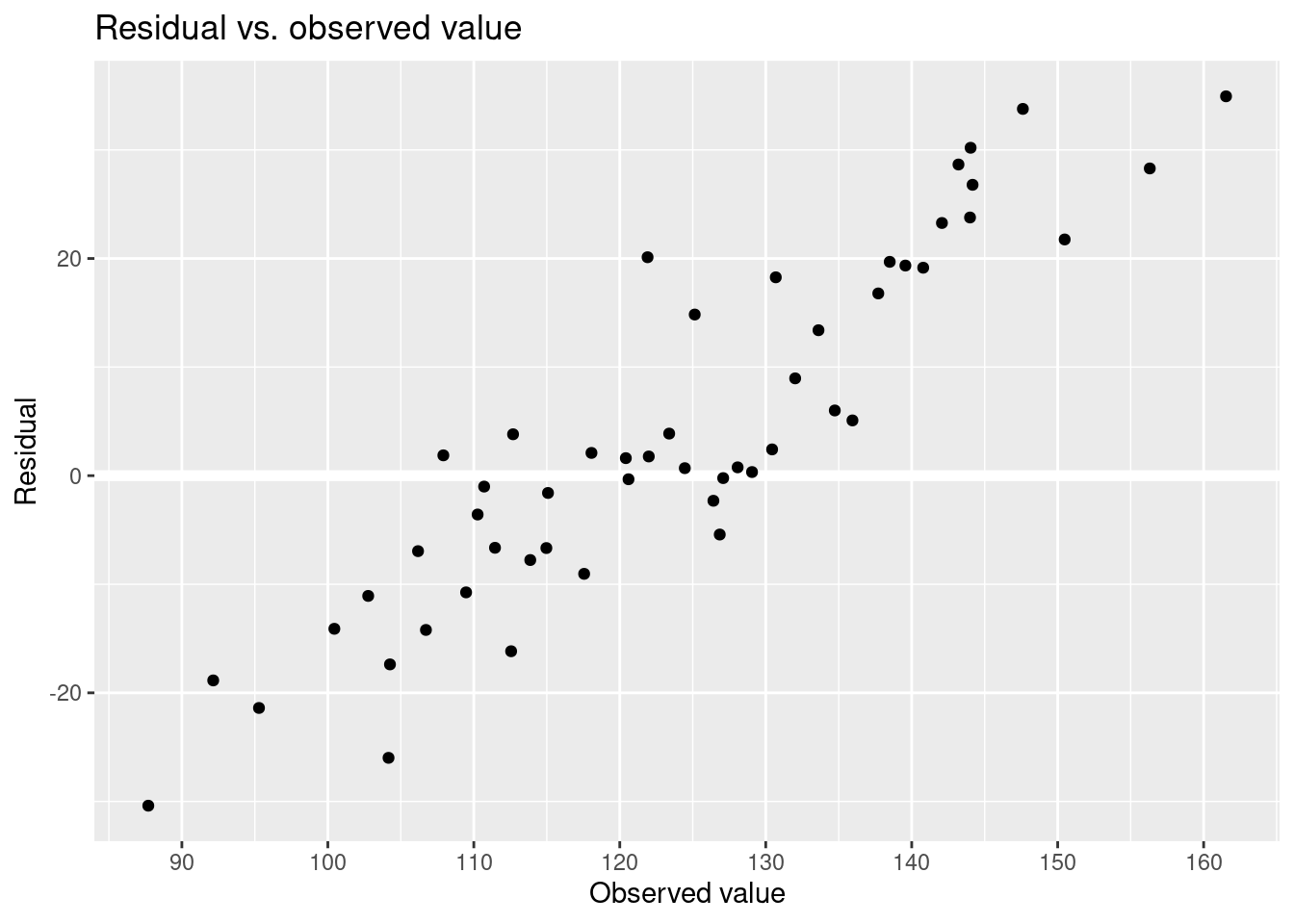
resid = final_sim - pred
)Residual vs. observed value.
Residual vs. predicted value.
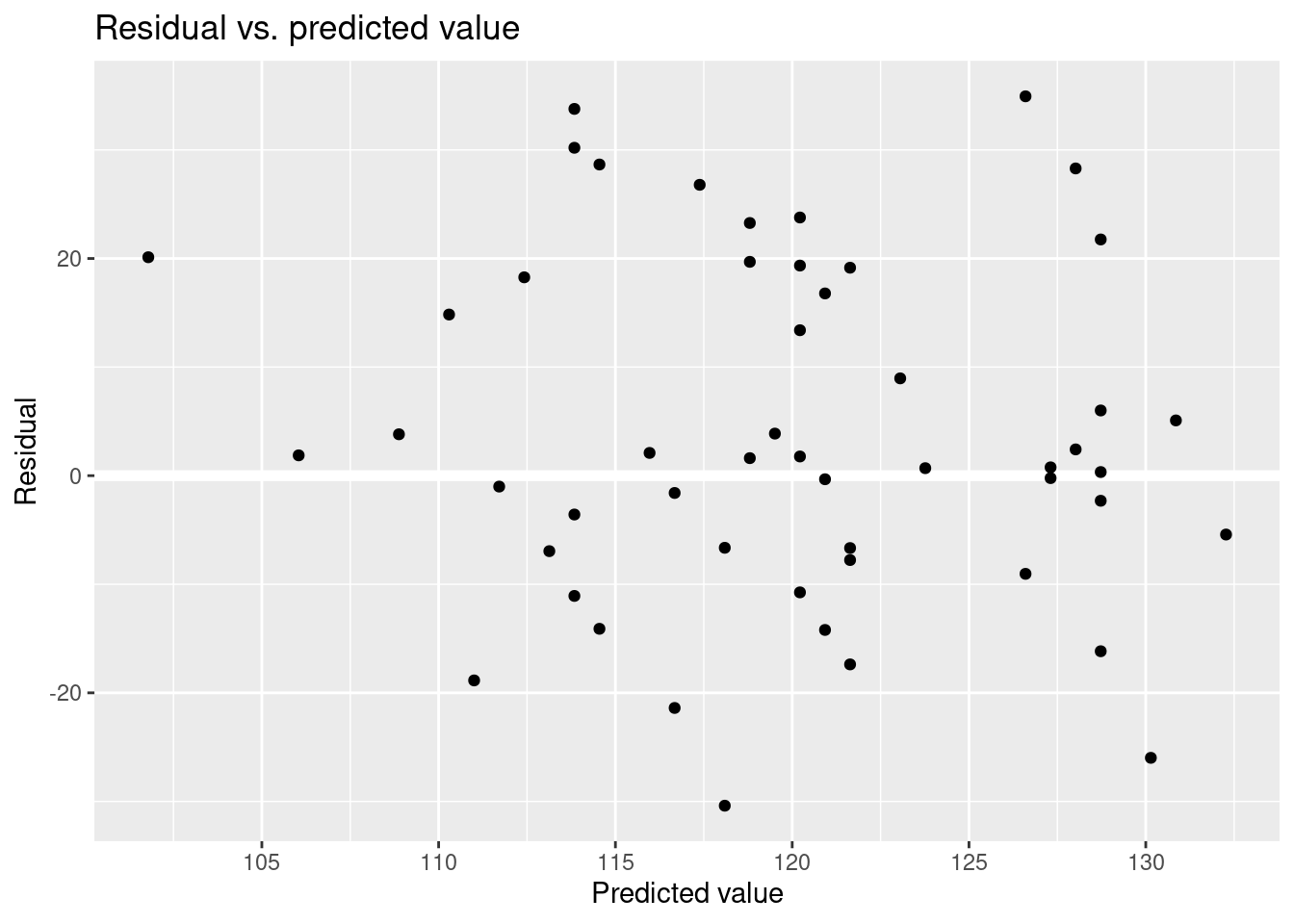
These are the type of plots you would see even if the model were correct.