Compare simulated with observed
Recall from previous chapter, we can more simply simulate using posterior_predict().
set.seed(970)
y_rep <- posterior_predict(fit)
# Each row of the matrix y_rep is 66 columns of simulated `newcomb` dataVerify these are the same thing (using same seed).
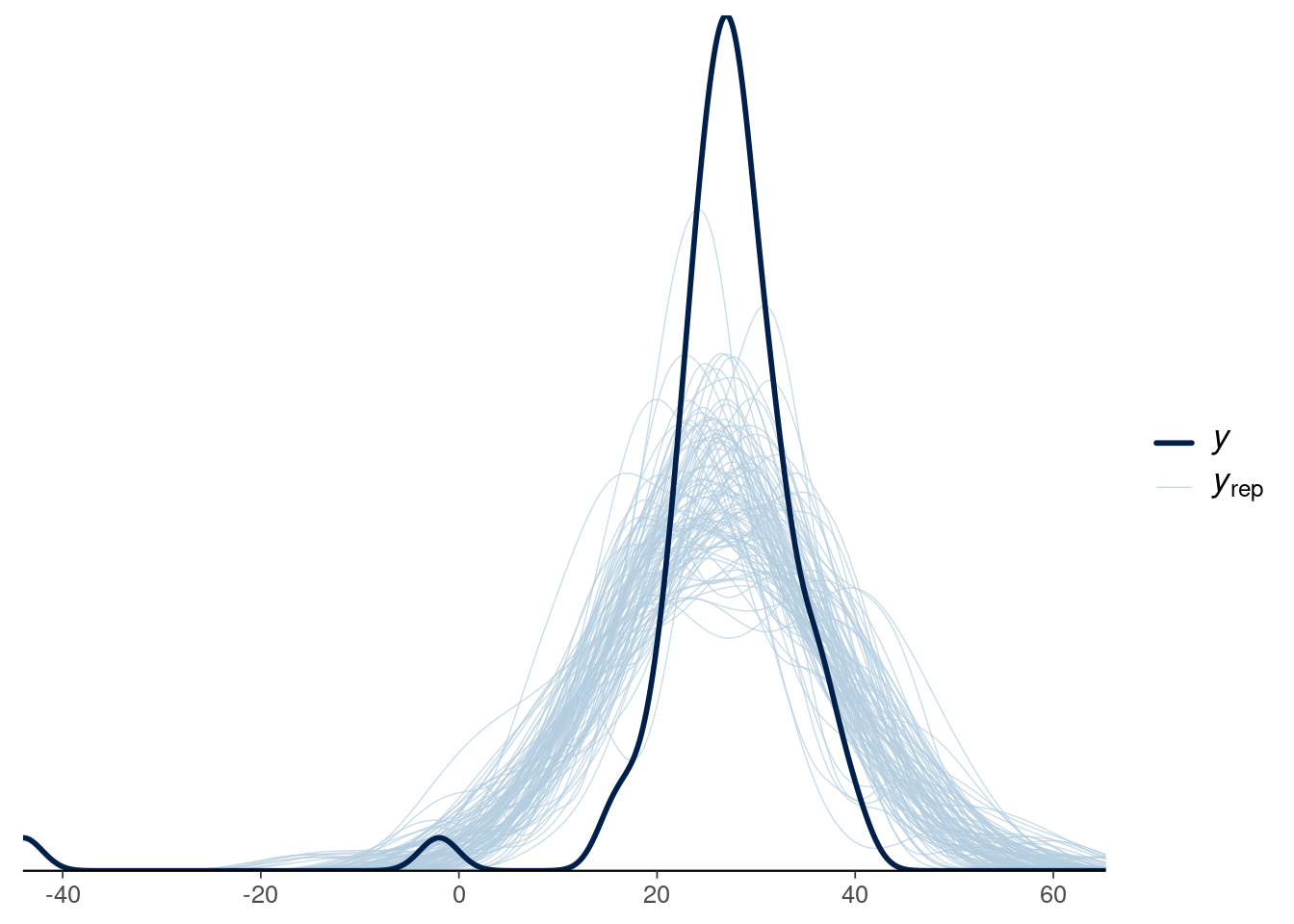
## [1] 0We can use bayesplot package to plot kernel density of data and n_rep sample replicates t.
set.seed(792)
n_rep <- 100
sims_sample <- sample(n_sims, n_rep)
ppc_dens_overlay(y = newcomb$y, yrep = y_rep[sims_sample, ]) +
theme(
axis.line.y = element_blank(),
text = element_text(family = "sans")
)
Checking model fit using a numerical data summary
Choose your own statistic! Here we choose the minimum measurement.
Plot test statistic for data and replicates using bayesplot.
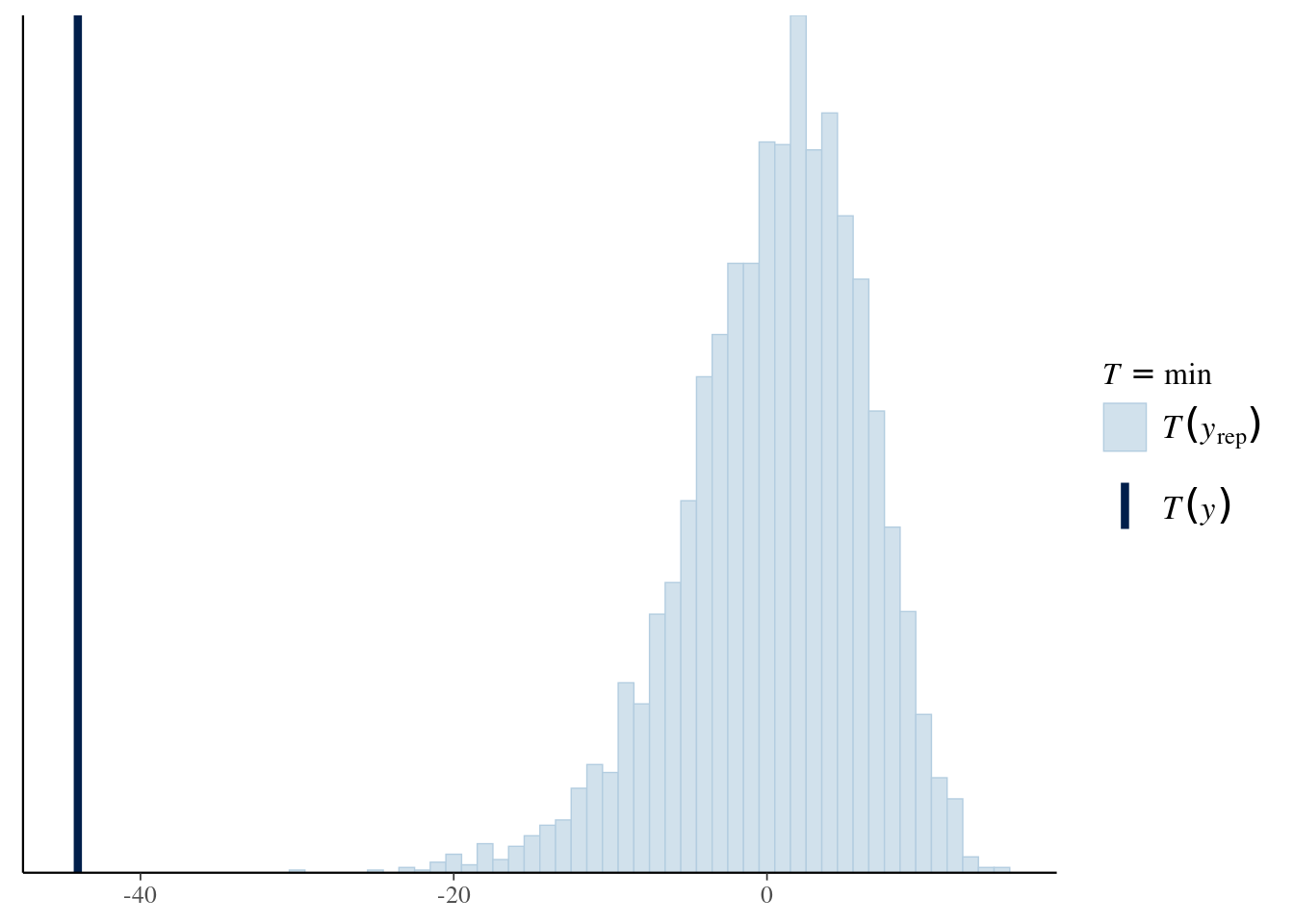
A normal model clearly doesn’t work, a revised model might use an asymmetric distribution or long tailed distribution in place of the normal.