8.1
Load libraries.
library(igraph)
library(tidyverse)We begin by reading in the data from the GSS network module 2004.
gss_url <-"https://raw.githubusercontent.com/mahoffman/stanford_networks/main/data/gss_local_nets.csv"
gss <- read_csv(gss_url) ## Rows: 1426 Columns: 41
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (13): sex, race, partyid, relig, educ2, educ3, educ4, educ5, relig1, rel...
## dbl (28): age, numgiven, close12, close13, close14, close15, close23, close2...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Let’s have a broad overview of the data
visdat::vis_dat(gss,sort_type = FALSE)+
labs(title = "GSS data",
subtitle = glue::glue("#rows = {nrow(gss)}, #columns = {ncol(gss)} "))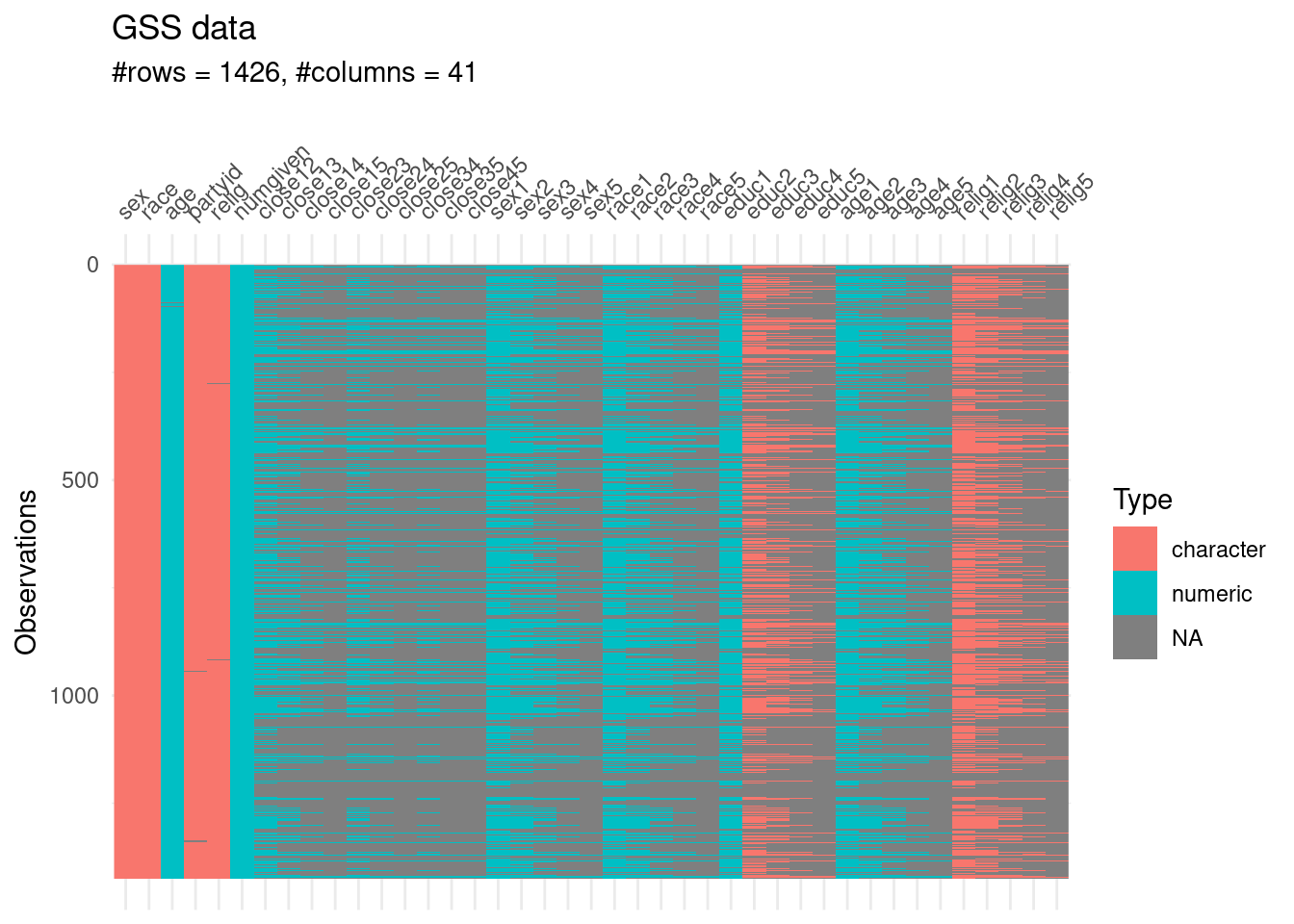
The first five concern the attributes of a given respondent:
sex
age
race
partyid
religion.
atrr_n <- 5
visdat::vis_dat(gss[,1:atrr_n],sort_type = FALSE)+
labs(title = "GSS data: attributes of a given respondent",
subtitle = glue::glue("#rows = {nrow(gss[,1:atrr_n])}, #columns = {ncol(gss[,1:atrr_n])} "))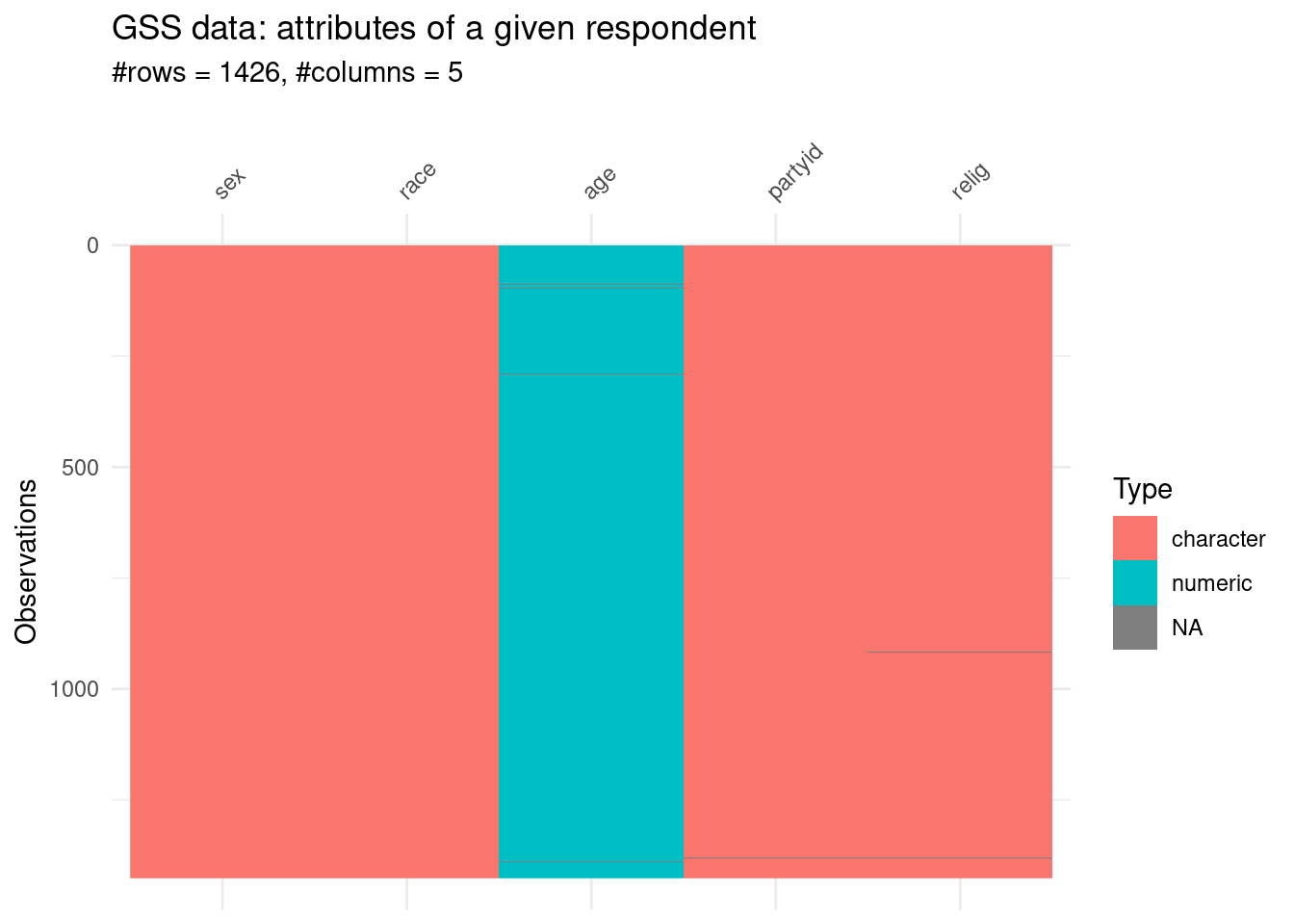
The basic idea of the module was to ask people about up to five others with whom they discussed “important matters” in the past six months. The respondents reported the number of people whom they discussed “important matters”:
numgiven: the number of others whom they repondents discussed important matters with.
“close” columns: The relationship between others (e.g., close12 is the closeness of person 1 to person 2, for each respondent).
“sex, race, age” columns: attributes of each of the others (n=5) in the ego network. (3*5)
net_n <- 41
visdat::vis_dat(gss[,(atrr_n+1):net_n],sort_type = FALSE)+
labs(title = "GSS data: ''netwok' part",
subtitle = glue::glue("#rows = {nrow(gss[,(atrr_n+1):net_n])}, #columns = {ncol(gss[,(atrr_n+1):net_n])} "))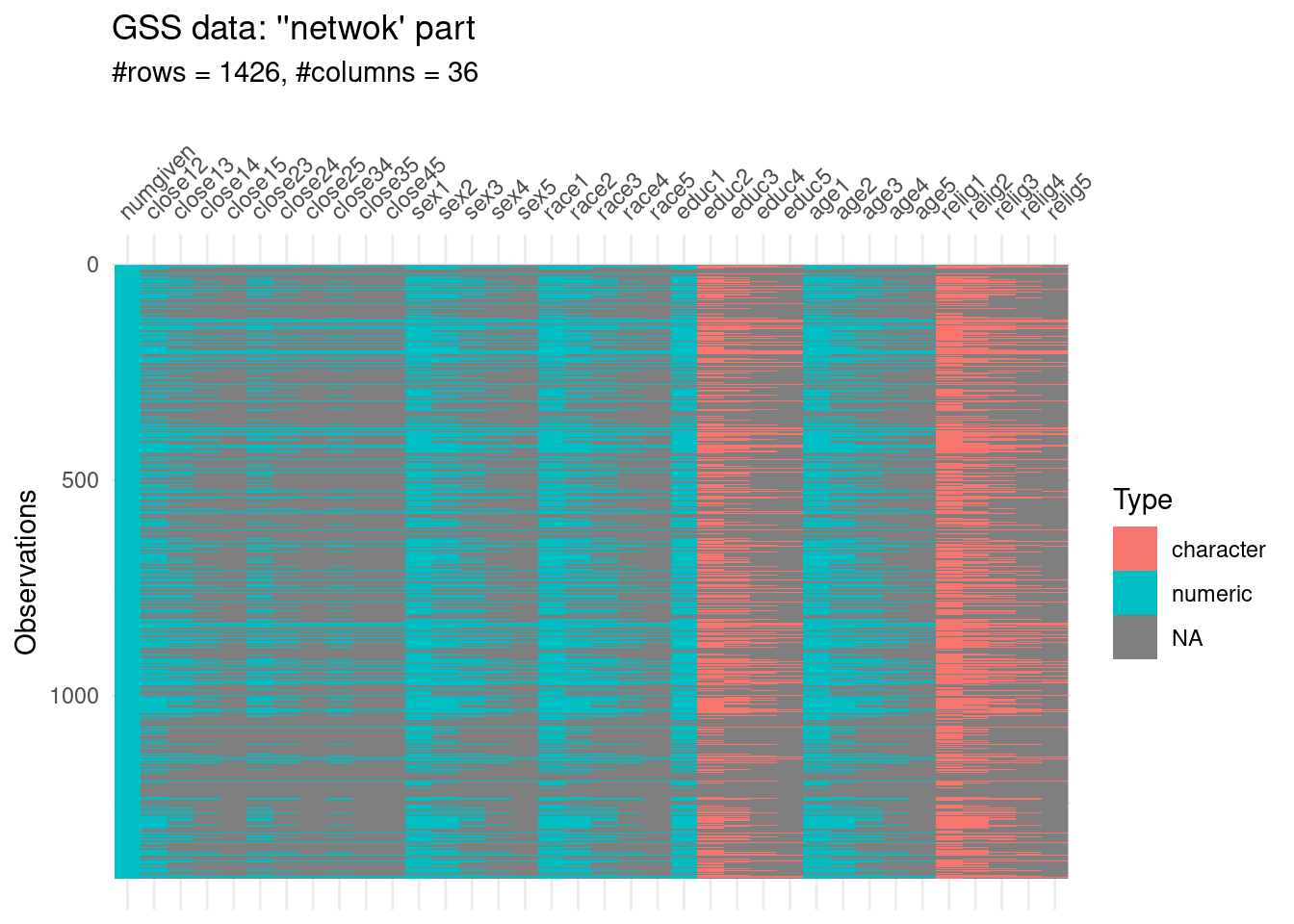
To do so, we have to first turn the variables close12 through close45 into an edge list, one for each respondent.
ties <- gss %>%
dplyr::select(starts_with("close"))
head(ties)## # A tibble: 6 × 10
## close12 close13 close14 close15 close23 close24 close25 close34 close35
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 NA NA NA NA NA NA NA NA NA
## 2 NA NA NA NA NA NA NA NA NA
## 3 1 2 0 NA 2 2 NA 1 NA
## 4 2 0 2 NA 2 2 NA 2 NA
## 5 NA NA NA NA NA NA NA NA NA
## 6 0 2 1 1 1 1 1 2 2
## # ℹ 1 more variable: close45 <dbl>A function, which uses the code above to turn any row in the ties data set into an ego network, and then apply that function to every row in the data
make_ego_nets <- function(tie){
# make the matrix
mat = matrix(nrow = 5, ncol = 5)
# assign the tie values to the lower triangle
mat[lower.tri(mat)] <- as.numeric(tie)
# symmetrize
mat[upper.tri(mat)] = t(mat)[upper.tri(mat)]
# identify missing values
na_vals <- is.na(mat)
# identify rows where all values are missing
non_missing_rows <- rowSums(na_vals) < nrow(mat)
# if any rows
if(sum(!non_missing_rows) > 0){
mat <- mat[non_missing_rows,non_missing_rows]
}
diag(mat) <- 0
ego_net <- graph.adjacency(mat, mode = "undirected", weighted = T)
return(ego_net)
}A simpler approach
make_ego_nets_simple <- function(tie){
#get the all possible links among others
tie <- tie %>% unlist
#remove missing links
tie <- tie[!is.na(tie)]
#remove zero links
tie <- tie[tie!=0]
#get the identity of linked pairs
others <- str_extract(names(tie), "[0-9]+")
#split the linked others
others_link <- str_split(others, "",simplify = TRUE) %>% as.data.frame
#make edge list of others
others_link <- cbind(others_link, tie)
#ego graph with
graph_from_data_frame(others_link,
directed=FALSE)
}Clean ties before creating the networks
#set zero links as missing links
ties[ties==0] <- NA
#repondents where all links among others are missing
others_missing <- rowSums(is.na(ties))==ncol(ties)
#remove any respondent that falls in any of the above
ties <- ties[!(others_missing),]Compare the two functions
ego_nets <- apply(ties,1,make_ego_nets)
ego_nets_simple <- apply(ties,1,make_ego_nets_simple)
plot(ego_nets[[1]])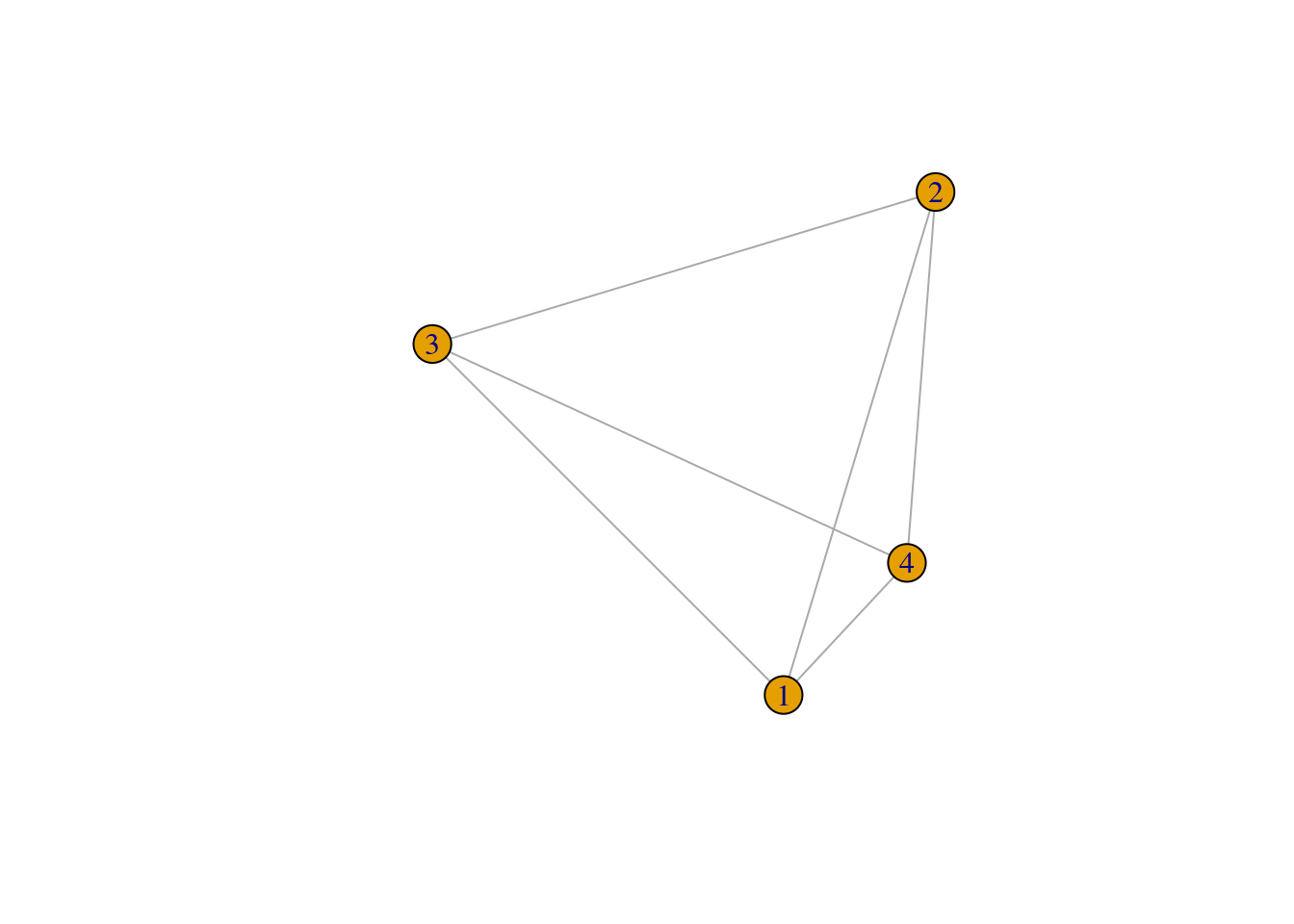
plot(ego_nets_simple[[1]])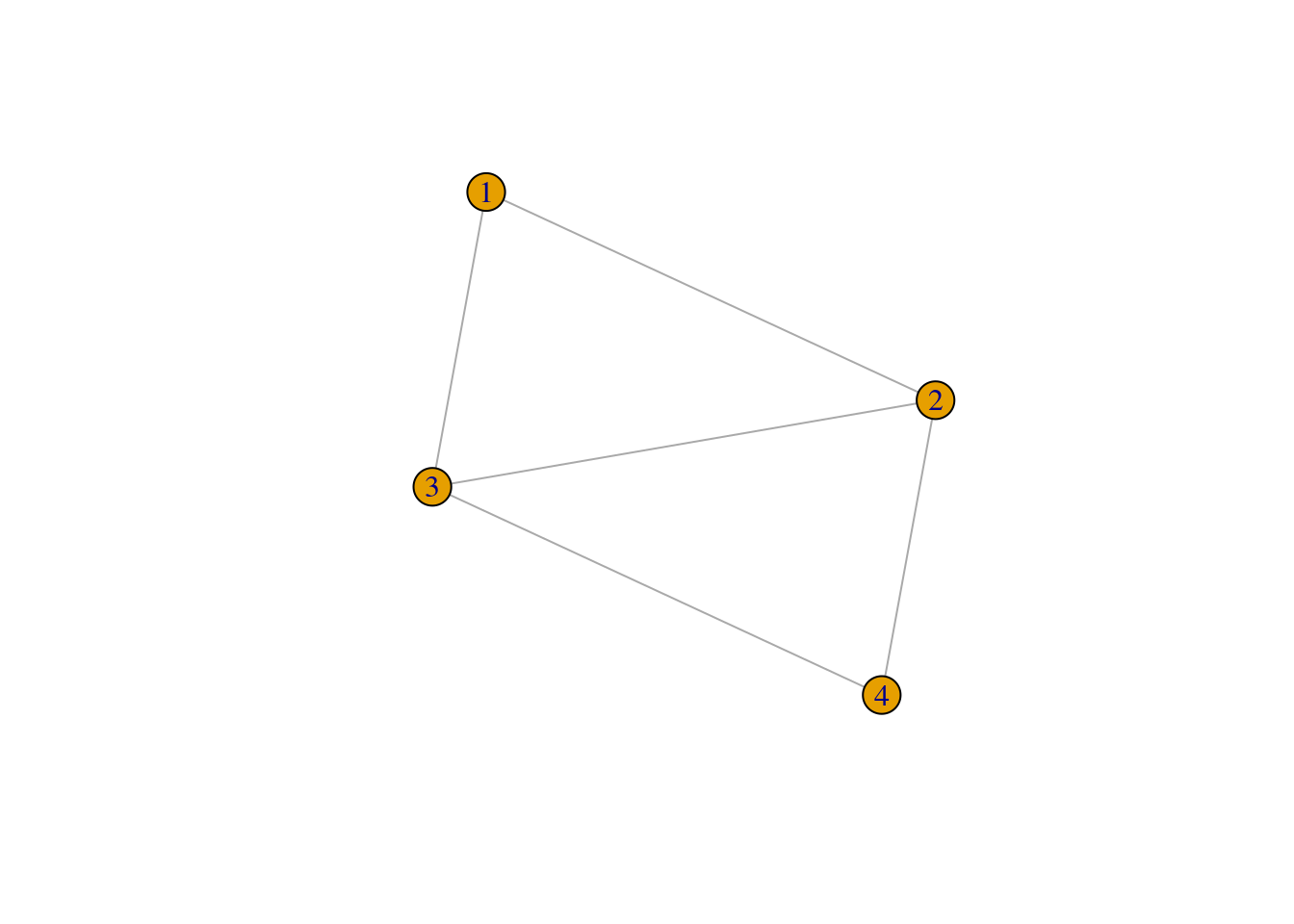
Not the same ! Where did thing go wrong?
ties[1,]## # A tibble: 1 × 10
## close12 close13 close14 close15 close23 close24 close25 close34 close35
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 2 NA NA 2 2 NA 1 NA
## # ℹ 1 more variable: close45 <dbl>