9.1 Load data
Regenerate data from previous chapter. (code below copied from previous chapter)
#read data
gss_url <-"https://raw.githubusercontent.com/mahoffman/stanford_networks/main/data/gss_local_nets.csv"
gss <- read_csv(gss_url) ## Rows: 1426 Columns: 41
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (13): sex, race, partyid, relig, educ2, educ3, educ4, educ5, relig1, rel...
## dbl (28): age, numgiven, close12, close13, close14, close15, close23, close2...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.#subset ties data
ties <- gss %>%
dplyr::select(starts_with("close"))
#set zero links as missing links
ties[ties==0] <- NA
#repondents where all links among others are missing
others_missing <- rowSums(is.na(ties))==ncol(ties)
#remove any respondent that falls in any of the above
ties <- ties[!(others_missing),]
#make a function to generate a network of ties
make_ego_nets_simple <- function(tie){
#get the all possible links among others
tie <- tie %>% unlist
#remove missing links
tie <- tie[!is.na(tie)]
#remove zero links
tie <- tie[tie!=0]
#get the identity of linked pairs
others <- str_extract(names(tie), "[0-9]+")
#split the linked others
others_link <- str_split(others, "",simplify = TRUE) %>% as.data.frame
#make edge list of others
others_link <- cbind(others_link, tie)
#ego graph with
graph_from_data_frame(others_link,
directed=FALSE)
}
#get all graphs
ego_nets_simple <- apply(ties,1,make_ego_nets_simple)
#
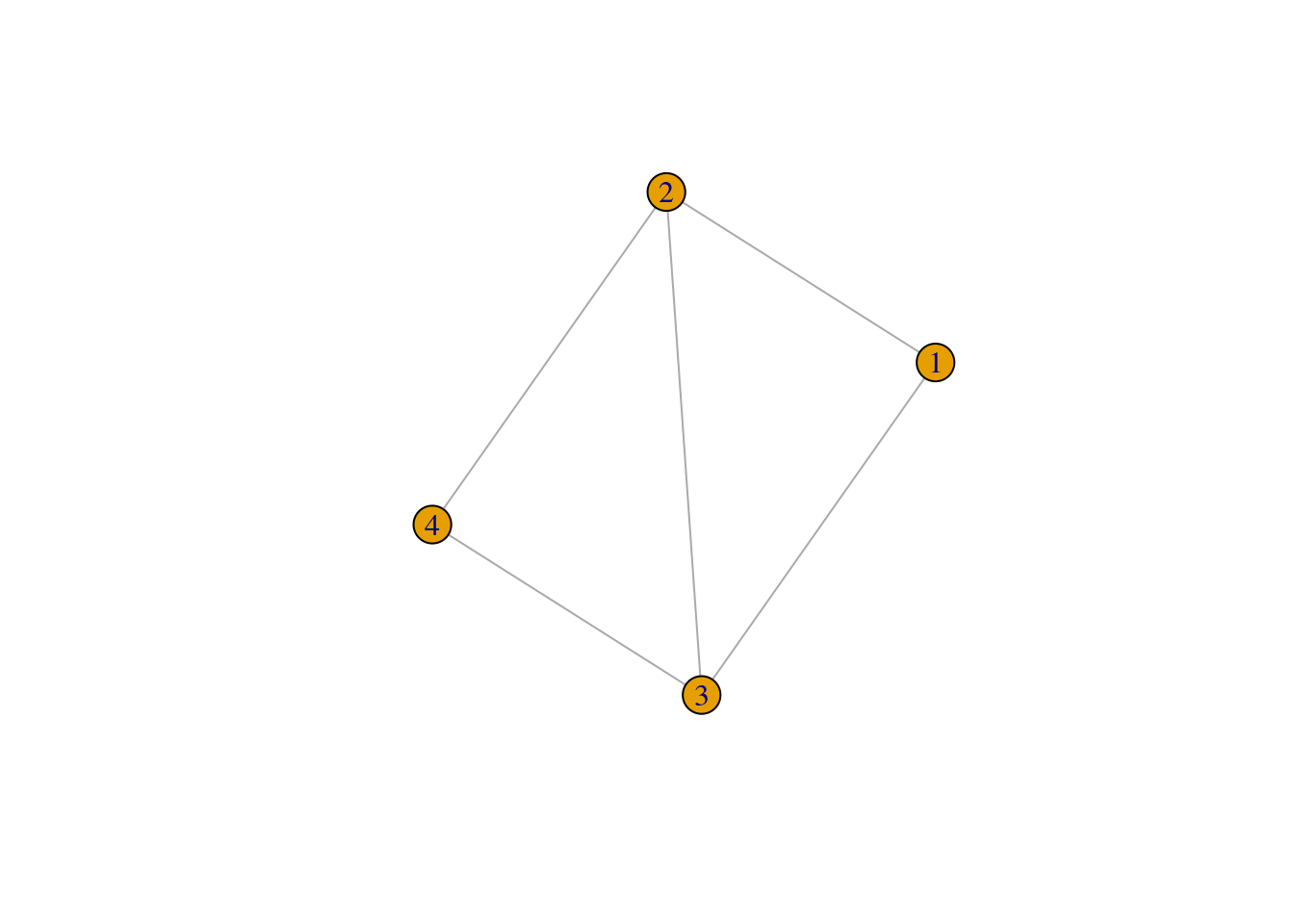
ties[1,]## # A tibble: 1 × 10
## close12 close13 close14 close15 close23 close24 close25 close34 close35
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 2 NA NA 2 2 NA 1 NA
## # ℹ 1 more variable: close45 <dbl>plot(ego_nets_simple[[1]])