6.4 From data to networks
suppressPackageStartupMessages(library(igraph))#edge list
personA <- c("Mark", "Mark", "Peter", "Peter", "Bob", "Jill")
personB <- c("Peter", "Jill", "Bob", "Aaron", "Jill", "Aaron")
edgelist <- cbind(PersonA = personA, PersonB = personB)
print(edgelist)## PersonA PersonB
## [1,] "Mark" "Peter"
## [2,] "Mark" "Jill"
## [3,] "Peter" "Bob"
## [4,] "Peter" "Aaron"
## [5,] "Bob" "Jill"
## [6,] "Jill" "Aaron"#adjacency matrix
adjacency <- matrix(c(0,1,0,1,0,1,0,1,0,1,0,1,0,1,0,1,0,1,0,1,0,1,0,1,0),
nrow = 5,
ncol = 5,
dimnames = list(c("Mark", "Peter", "Bob", "Jill", "Aaron"),
c("Mark", "Peter", "Bob", "Jill", "Aaron")))
print(adjacency)## Mark Peter Bob Jill Aaron
## Mark 0 1 0 1 0
## Peter 1 0 1 0 1
## Bob 0 1 0 1 0
## Jill 1 0 1 0 1
## Aaron 0 1 0 1 0Edge list into a network object.
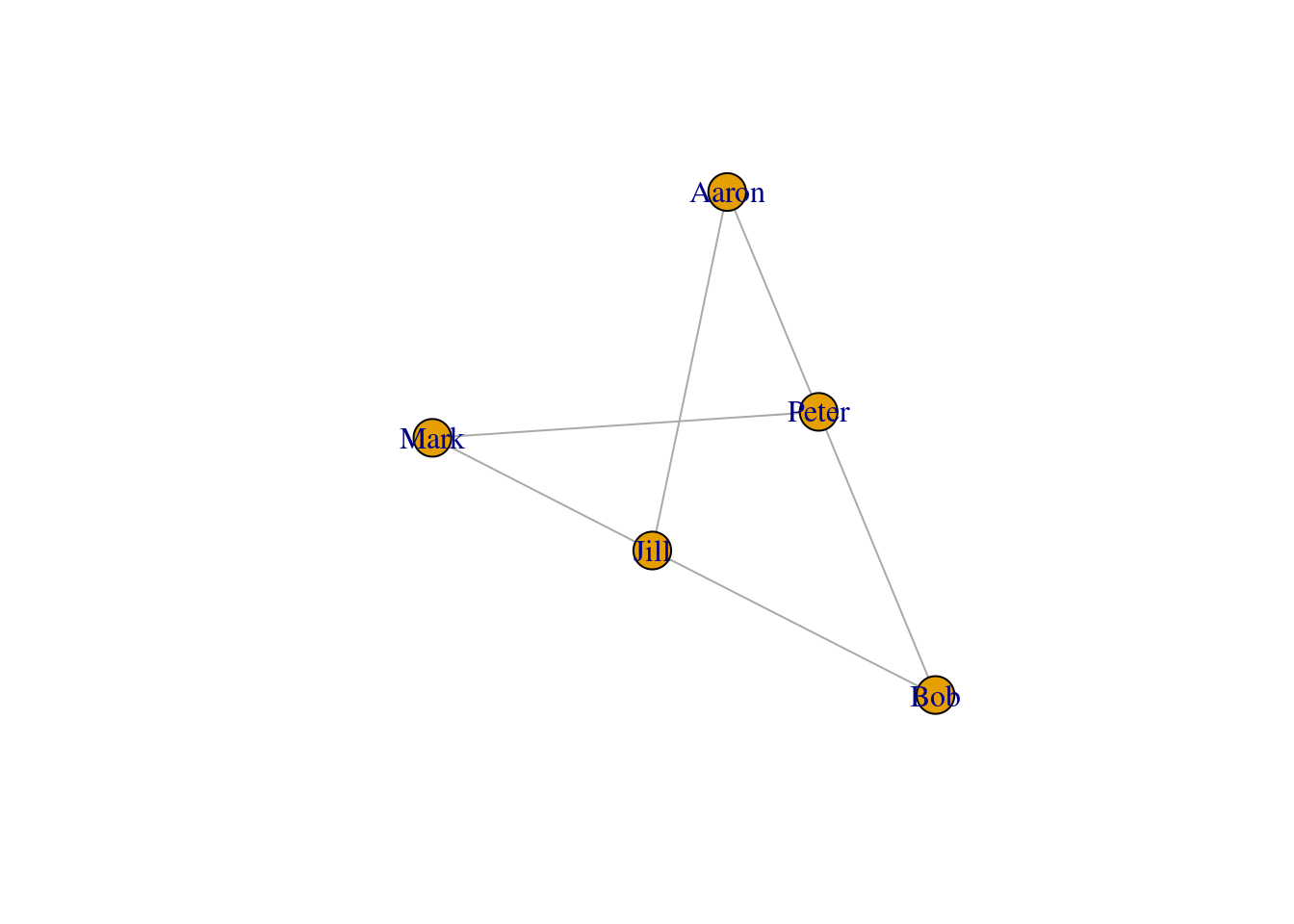
network <- graph.edgelist(edgelist, directed=FALSE)
network## IGRAPH 3e8ebd5 UN-- 5 6 --
## + attr: name (v/c)
## + edges from 3e8ebd5 (vertex names):
## [1] Mark --Peter Mark --Jill Peter--Bob Peter--Aaron Jill --Bob
## [6] Jill --AaronAdjacency matrix to network object.
network_adj <-graph.adjacency(adjacency,mode = "undirected" )UN: undirectedDN: directed- #vertices #edges
Vertices of network
V(network)## + 5/5 vertices, named, from 3e8ebd5:
## [1] Mark Peter Jill Bob AaronEdges of network
E(network)## + 6/6 edges from 3e8ebd5 (vertex names):
## [1] Mark --Peter Mark --Jill Peter--Bob Peter--Aaron Jill --Bob
## [6] Jill --AaronWe can visualize the network using the plot() function.
plot(network)