start_time <- Sys.time()
# using OLS to guide creation of prior distribution
OLS_model <- lm(bill_length_mm ~ flipper_length_mm,
data = penguin_class_df)
OLS_intercept <- coef(OLS_model)[1]
OLS_slope <- coef(OLS_model)[2]
mu_x <- mean(penguin_class_df$flipper_length_mm)
mu_y <- mean(penguin_class_df$bill_length_mm)
var_x <- var(penguin_class_df$flipper_length_mm)
var_y <- var(penguin_class_df$bill_length_mm)
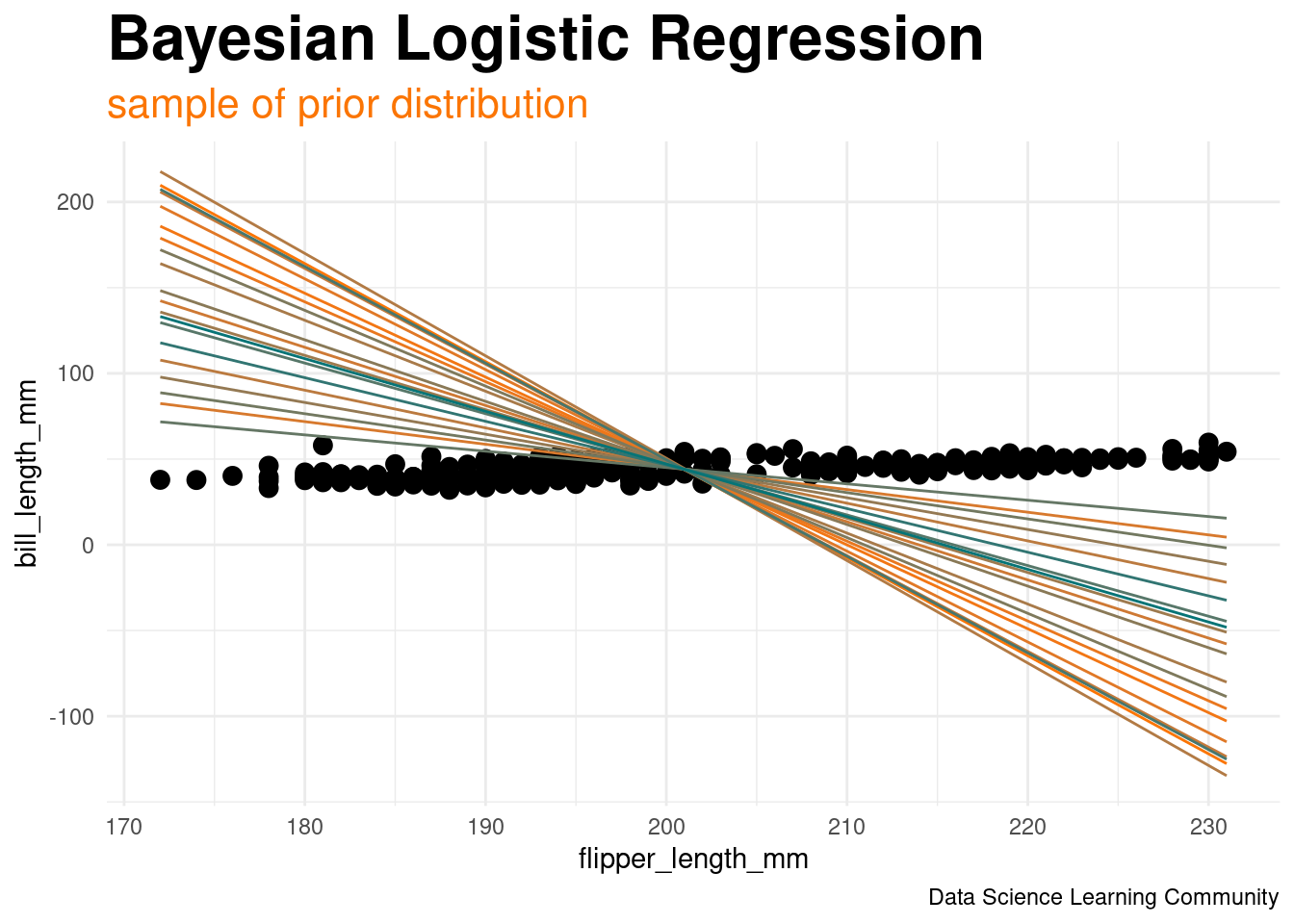
Bayesian_model_prior <- rstanarm::stan_glm(
formula = chinstrap_bool ~ flipper_length_mm + bill_length_mm,
data = penguin_class_df,
family = binomial,
prior_intercept = normal(OLS_intercept, 1),
prior = normal(c(mu_x, mu_y), c(var_x, var_y)),
prior_aux = exponential(1),
prior_PD = TRUE,
chains = 4, iter = 5000*2, refresh = 0, seed = 320)
Bayesian_model_post <- update(Bayesian_model_prior, prior_PD = FALSE)
end_time <- Sys.time()
print(end_time - start_time)
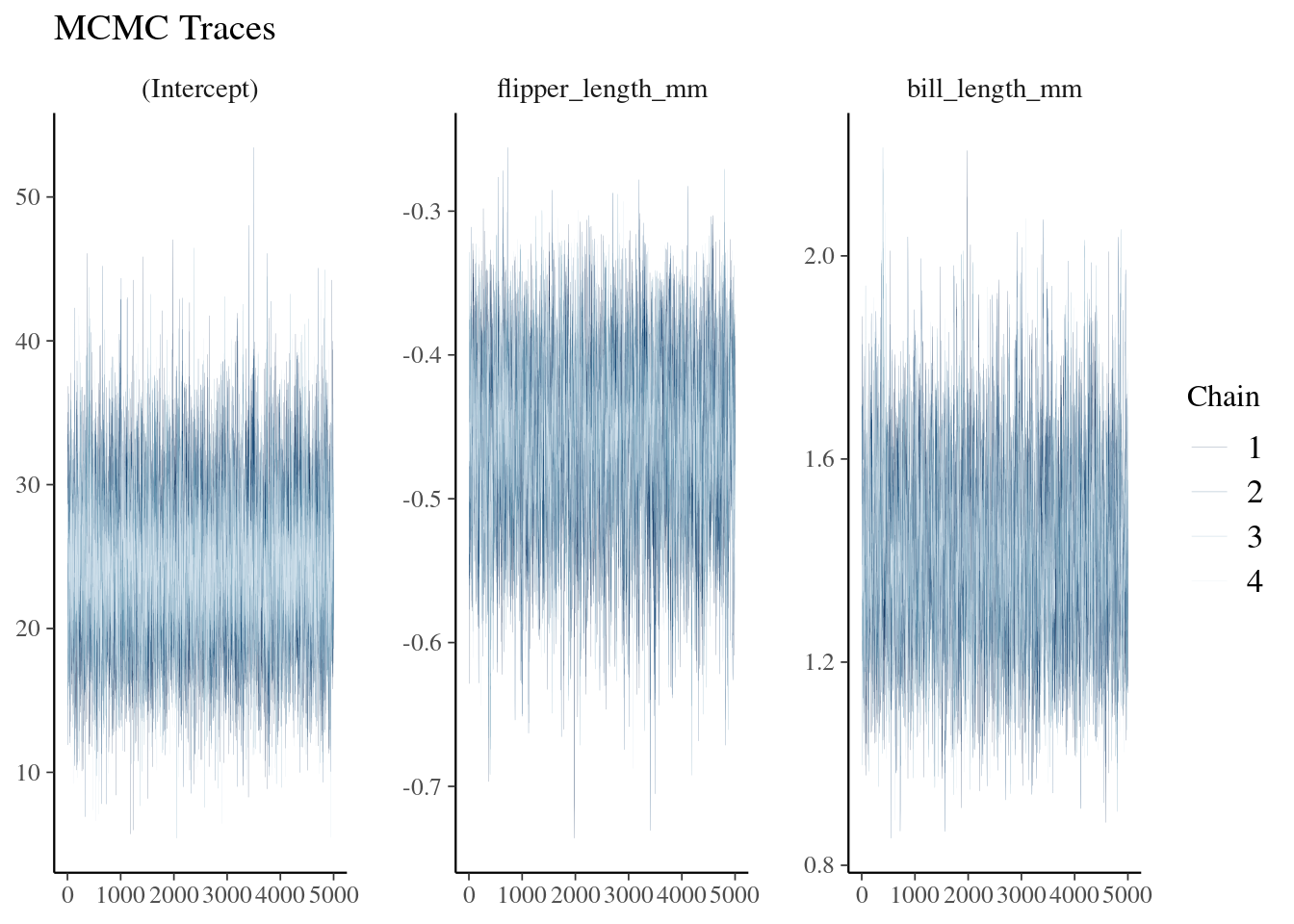
Posterior Distribution
bayesplot::mcmc_trace(Bayesian_model_post, size = 0.1) +
labs(title = "MCMC Traces")
R code
sim_df_post <- as.data.frame(Bayesian_model_post) |>
mutate(boundary_intercept = -1.0*`(Intercept)` / bill_length_mm,
boundary_slope = -1.0*flipper_length_mm / bill_length_mm)
# make data frame of line segments
sim_df_post_sample <- sim_df_post |>
mutate(x1 = flipper_min,
x2 = flipper_max,
y1 = boundary_intercept + boundary_slope * x1,
y2 = boundary_intercept + boundary_slope * x2) |>
slice_sample(n = 20) |>
mutate(line_id = 1:20)
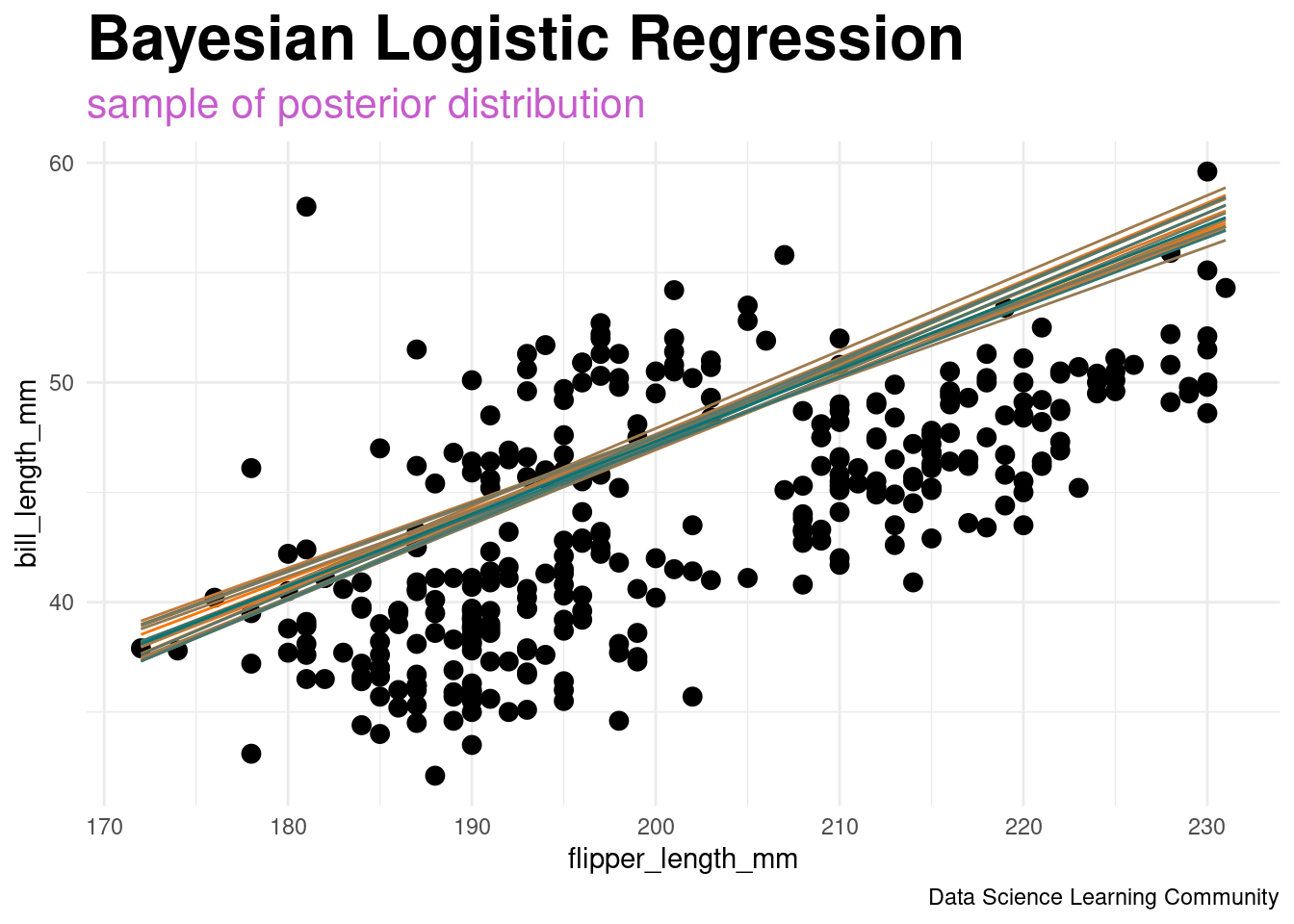
penguin_class_df |>
ggplot(aes(x = flipper_length_mm, y = bill_length_mm)) +
geom_point(size = 3) +
geom_segment(aes(x = x1, y = y1, xend = x2, yend = y2,
color = line_id, group = line_id),
data = sim_df_post_sample) +
labs(title = "Bayesian Logistic Regression",
subtitle = "<span style = 'color:#c65ccc'>sample of posterior distribution</span>",
caption = "Data Science Learning Community") +
scale_color_gradient(low = adelie_color, high = gentoo_color) +
theme_minimal() +
theme(legend.position = "none",
plot.title = element_markdown(face = "bold", size = 24),
plot.subtitle = element_markdown(size = 16))
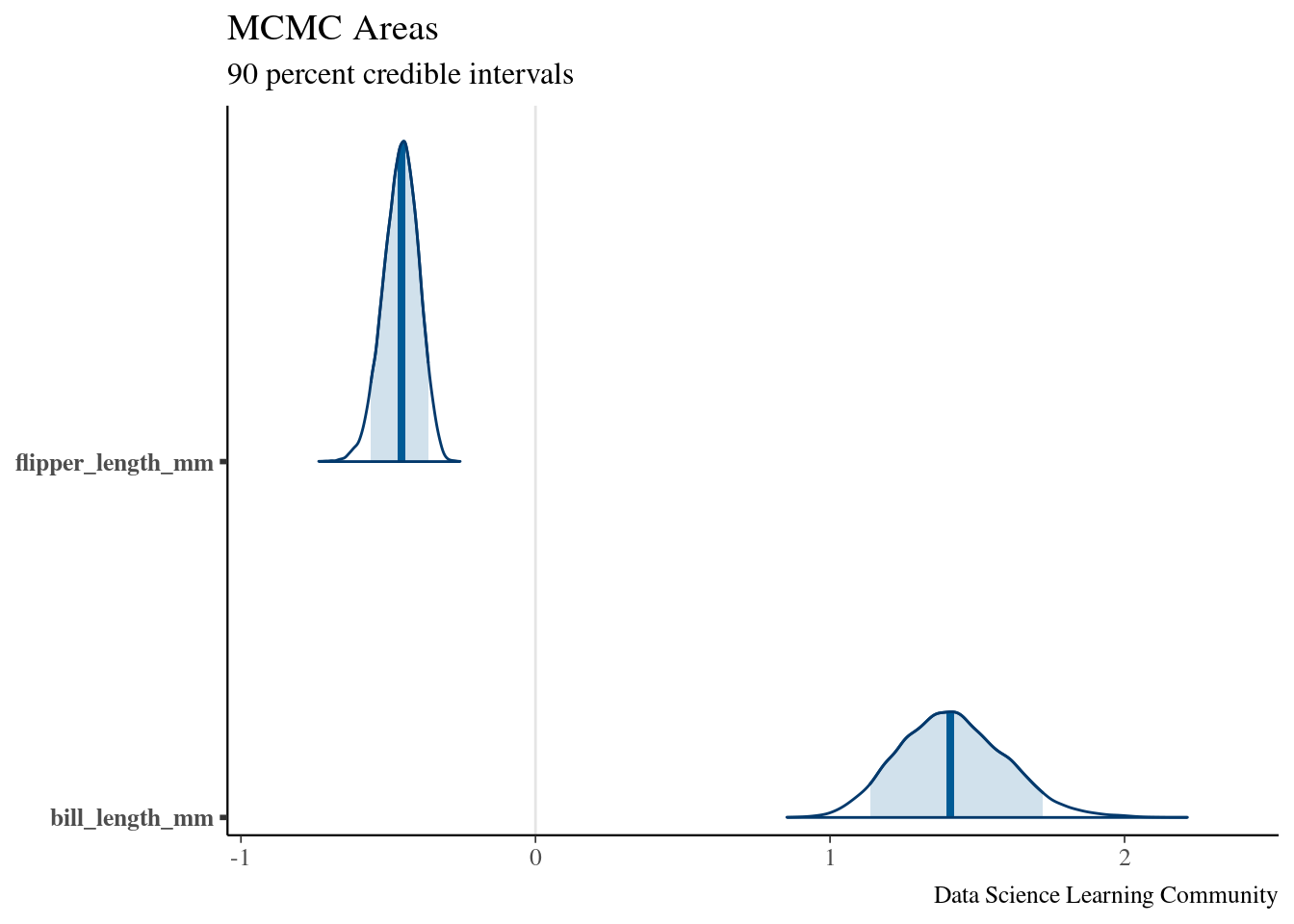
broom.mixed::tidy(Bayesian_model_post,
conf.int = TRUE, conf.level = 0.90) |>
mutate_if(is.numeric, round, digits = 4)
## # A tibble: 3 × 5
## term estimate std.error conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 24.2 5.39 16.0 33.6
## 2 flipper_length_mm -0.455 0.0599 -0.559 -0.364
## 3 bill_length_mm 1.41 0.182 1.14 1.72
R code
bayesplot::mcmc_areas(Bayesian_model_post,
pars = c("flipper_length_mm", "bill_length_mm"),
prob = 0.9) +
labs(title = "MCMC Areas",
subtitle = "90 percent credible intervals",
caption = "Data Science Learning Community")