Logistic Regression in R
Palmer Penguins Example
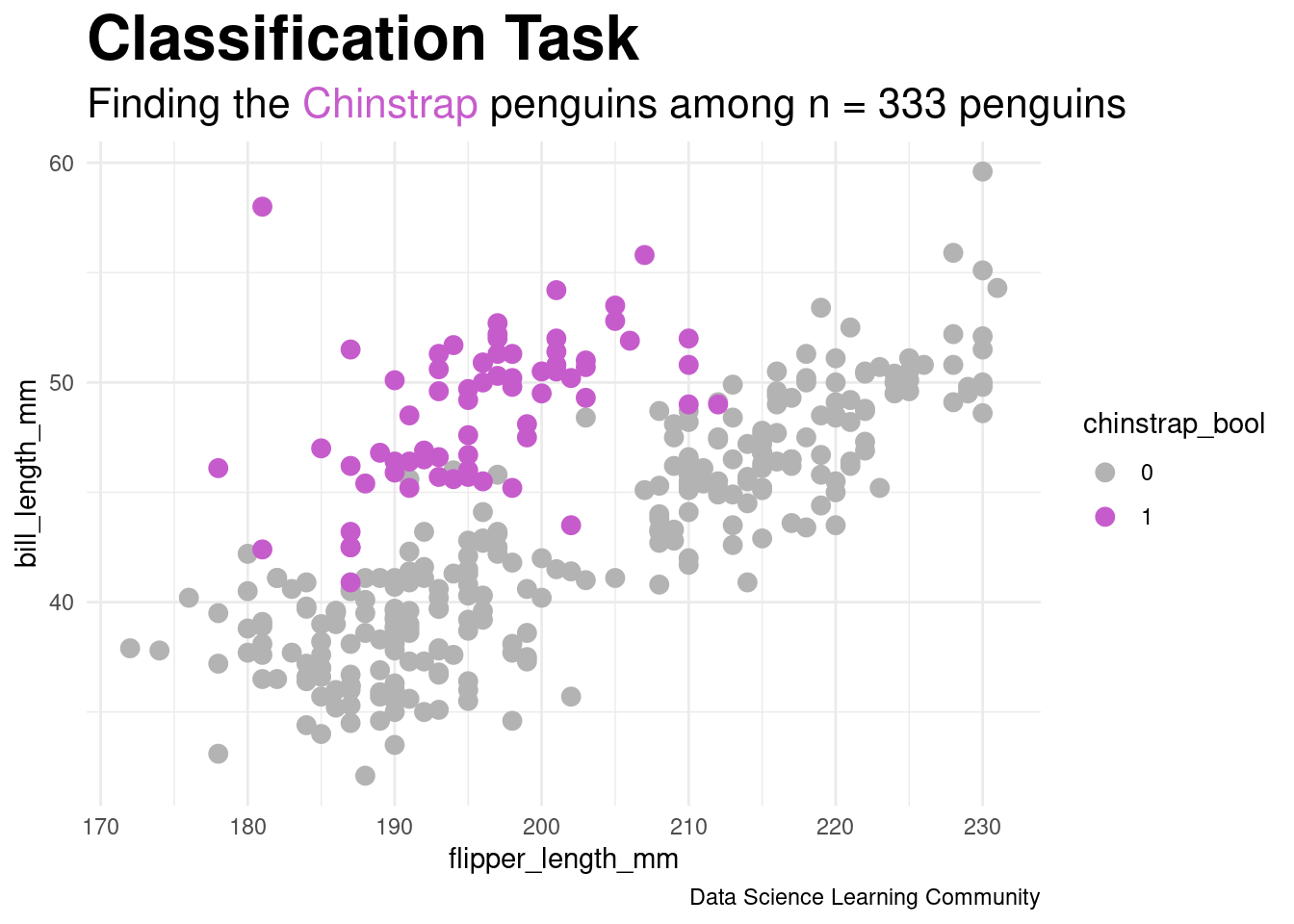
R Code
adelie_color = "#fb7504"
chinstrap_color = "#c65ccc"
gentoo_color = "#067476"
penguin_class_df <- penguins |>
na.omit() |>
mutate(chinstrap_bool = ifelse(species == "Chinstrap", 1, 0)) |>
mutate(across(chinstrap_bool, as.factor)) #https://stackoverflow.com/questions/33180058/coerce-multiple-columns-to-factors-at-once
penguin_class_df |>
ggplot(aes(x = flipper_length_mm, y = bill_length_mm,
color = chinstrap_bool)) +
geom_point(size = 3) +
labs(title = "Classification Task",
subtitle = "Finding the <span style = 'color:#c65ccc'>Chinstrap</span> penguins among n = 333 penguins",
caption = "Data Science Learning Community") +
scale_color_manual(values = c("gray70", chinstrap_color)) +
theme_minimal() +
theme(plot.title = element_markdown(face = "bold", size = 24),
plot.subtitle = element_markdown(size = 16))Generalized Linear Models
logistic_model <- stats::glm(chinstrap_bool ~ flipper_length_mm + bill_length_mm,
data = penguin_class_df,
family = binomial) #makes logistic regression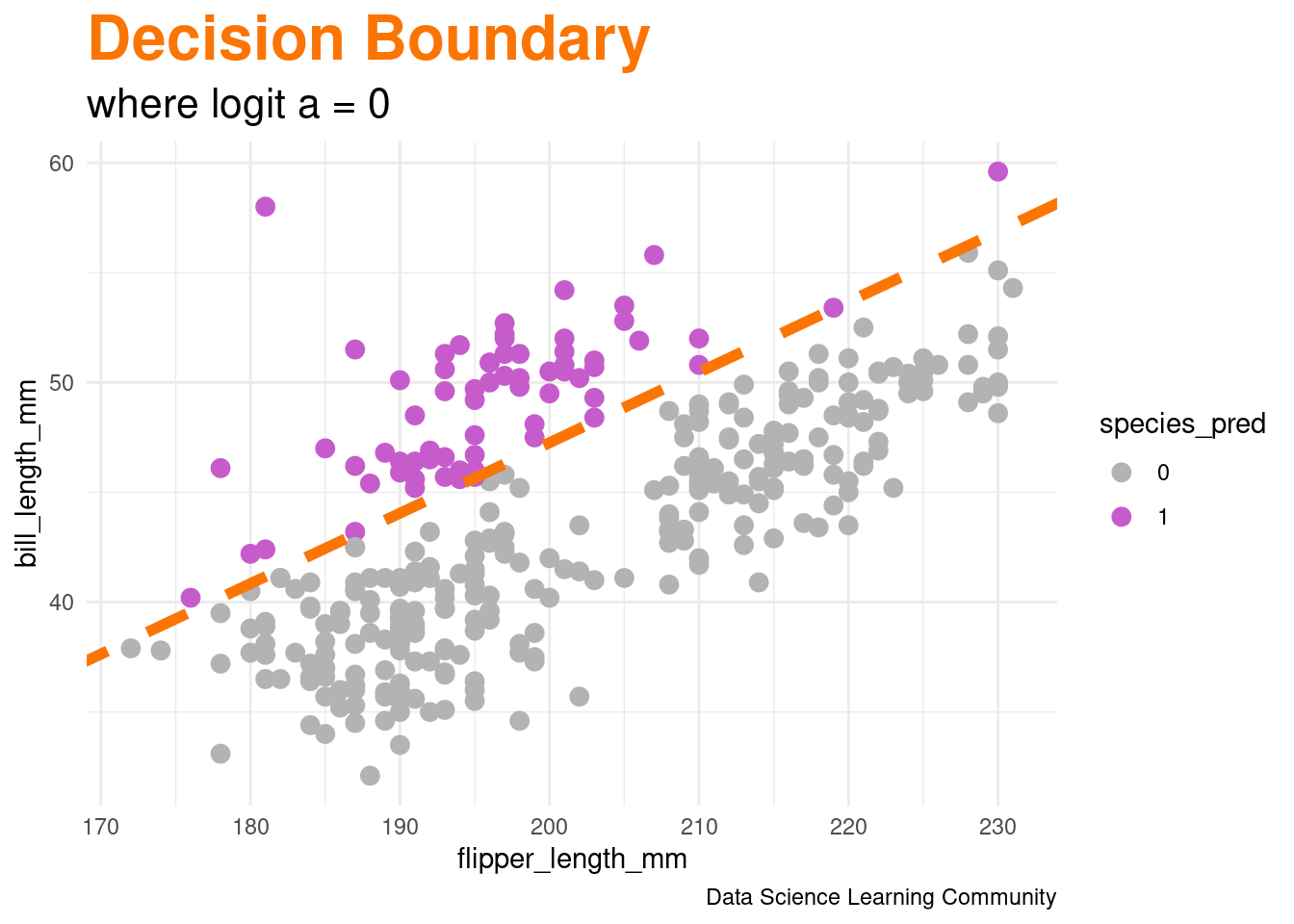
R code
# https://stats.stackexchange.com/questions/6206/how-to-plot-decision-boundary-in-r-for-logistic-regression-model
beta_0 <- coef(logistic_model)[1]
beta_1 <- coef(logistic_model)[2]
beta_2 <- coef(logistic_model)[3]
boundary_slope <- -1.0 * beta_1 / beta_2
boundary_intercept <- -1.0 * beta_0 / beta_2
penguin_pred_df <- penguin_class_df |>
mutate(species_pred = ifelse(
bill_length_mm > boundary_intercept + boundary_slope * flipper_length_mm,
1,0)) |>
mutate(across(species_pred, as.factor))
penguin_pred_df |>
ggplot(aes(x = flipper_length_mm, y = bill_length_mm,
color = species_pred)) +
geom_point(size = 3) +
geom_abline(intercept = boundary_intercept,
slope = boundary_slope,
color = adelie_color,
linewidth = 2,
linetype = 2) +
labs(title = "<span style = 'color:#fb7504'>Decision Boundary</span>",
subtitle = "where logit a = 0",
caption = "Data Science Learning Community") +
scale_color_manual(values = c("gray70", chinstrap_color)) +
theme_minimal() +
theme(plot.title = element_markdown(face = "bold", size = 24),
plot.subtitle = element_markdown(size = 16))penguin_pred_df |>
janitor::tabyl(chinstrap_bool, species_pred) |>
janitor::adorn_totals(c("row", "col"))## chinstrap_bool 0 1 Total
## 0 258 7 265
## 1 8 60 68
## Total 266 67 333- accuracy: 0.9550
- sensitivity: 0.8824
- specificity: 0.9736