Measuring performance
Learning objectives:
- Understand how to improve your code for making it faster
- Learn what are the tools for improving your code
- Test how to profile your code
Introduction
“Before you can make your code faster, you first need to figure out what’s making it slow.”

SLOW DOWN TO LEARN HOW TO CODE FASTER | credits: packtpub.com
- profile your code: measure the run-time of each line of code using realistic inputs
- experiment with alternatives to find faster code
- microbenchmark to measure the difference in performance.
Profiling
The tool to use is a profiler, it allows for sampling the code performance through stopping the execution of code every few milliseconds and recording all the steps.
Example:
Profile the execution of f():
profvis::pause() is used instead of Sys.sleep()
profile f(), with utils::Rprof()#> NULL#> sample.interval=100000
#> "pause" "f" "eval" "eval" "withVisible" "withCallingHandlers" "eval" "eval" "with_handlers" "doWithOneRestart" "withOneRestart" "withRestartList" "doWithOneRestart" "withOneRestart" "withRestartList" "withRestarts" "evaluate::evaluate" "evaluate" "in_dir" "in_input_dir" "eng_r" "block_exec" "call_block" "process_group" "withCallingHandlers" "xfun:::handle_error" "process_file" "knitr::knit" "rmarkdown::render" "execute" ".main"
#> "pause" "g" "f" "eval" "eval" "withVisible" "withCallingHandlers" "eval" "eval" "with_handlers" "doWithOneRestart" "withOneRestart" "withRestartList" "doWithOneRestart" "withOneRestart" "withRestartList" "withRestarts" "evaluate::evaluate" "evaluate" "in_dir" "in_input_dir" "eng_r" "block_exec" "call_block" "process_group" "withCallingHandlers" "xfun:::handle_error" "process_file" "knitr::knit" "rmarkdown::render" "execute" ".main"
#> "pause" "h" "g" "f" "eval" "eval" "withVisible" "withCallingHandlers" "eval" "eval" "with_handlers" "doWithOneRestart" "withOneRestart" "withRestartList" "doWithOneRestart" "withOneRestart" "withRestartList" "withRestarts" "evaluate::evaluate" "evaluate" "in_dir" "in_input_dir" "eng_r" "block_exec" "call_block" "process_group" "withCallingHandlers" "xfun:::handle_error" "process_file" "knitr::knit" "rmarkdown::render" "execute" ".main"
#> "pause" "h" "f" "eval" "eval" "withVisible" "withCallingHandlers" "eval" "eval" "with_handlers" "doWithOneRestart" "withOneRestart" "withRestartList" "doWithOneRestart" "withOneRestart" "withRestartList" "withRestarts" "evaluate::evaluate" "evaluate" "in_dir" "in_input_dir" "eng_r" "block_exec" "call_block" "process_group" "withCallingHandlers" "xfun:::handle_error" "process_file" "knitr::knit" "rmarkdown::render" "execute" ".main"Visualising profiles
Makes easier to build up a mental model of what you need to change:
profvis::profvis()
utils::summaryRprof()Memory profiling and the garbage collector
Profiling a loop that modifies an existing variable:
You can figure out what is the source of the problem by looking at the memory column. In this case, copy-on-modify acts in each iteration of the loop creating another copy of x.
Limitations
- Profiling does not extend to C code
- Anonymous functions are hard to figure out
- Arguments are evaluated inside another function
Exercise
?rm()Microbenchmarking
Measurement of the performance of a very small piece of code is useful for comparing small snippets of code for specific tasks.

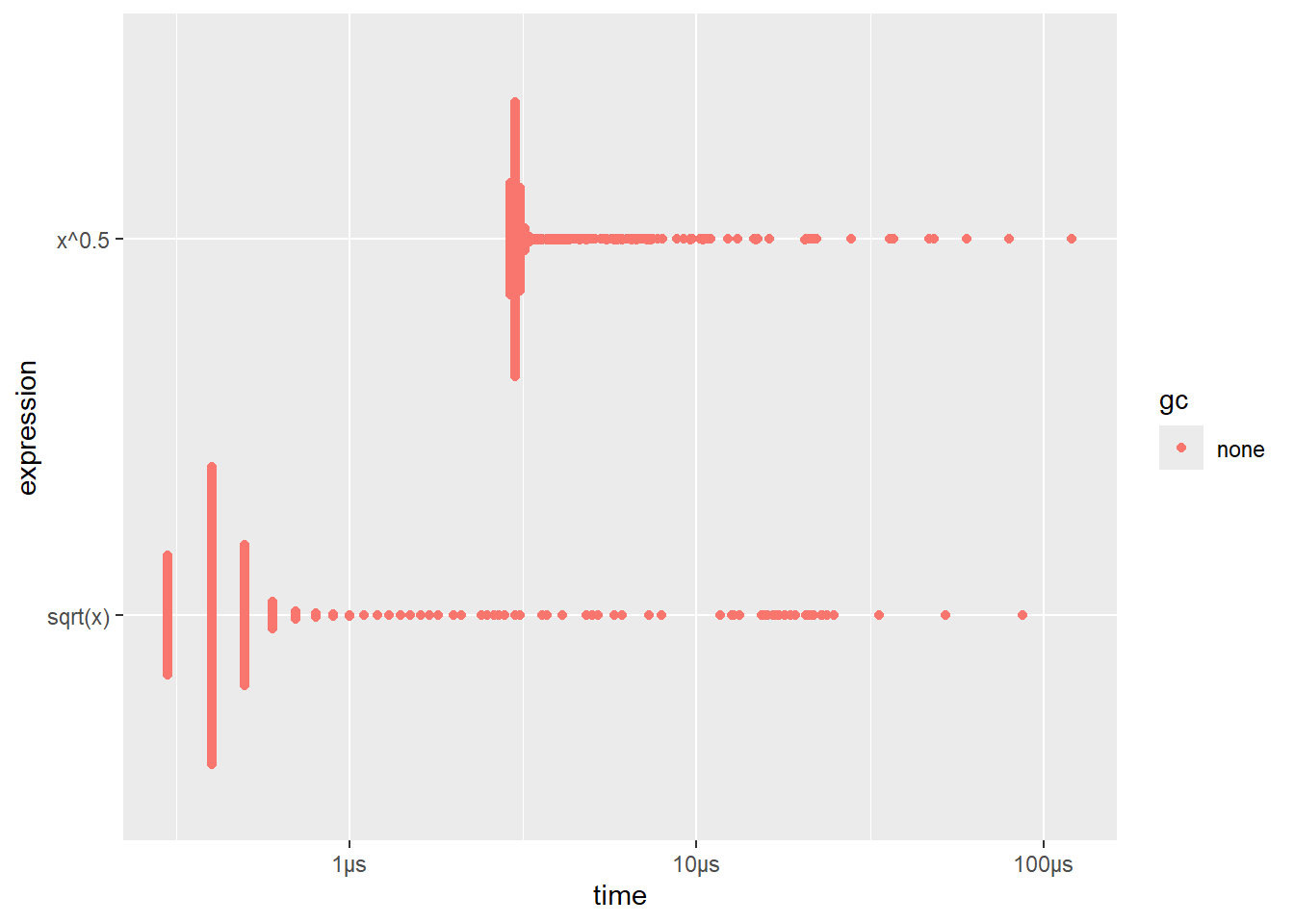
The {bench} package uses a high precision time.
bench::mark()#> # A tibble: 2 × 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 sqrt(x) 300ns 400ns 1977692. 848B 0
#> 2 x^0.5 2.9µs 3µs 321570. 848B 0- heavily right-skewed distribution