class: center, middle, inverse, title-slide # Advanced R: Chapters 23 & 24 ## All about performance ### Jon Leslie (<span class="citation">@jlesliedata</span>) ### Last Updated: 2020-09-02 --- <style> .pull-more-left { float: left; width: 40%; } .pull-less-right { float: right; width: 56%; } .pull-less-right ~ * { clear: both; } .remark-code-line { font-size: 0.7em !important; } </style> # How to go fast - Find out what's making it slow (Chapter 23) - Experiment with faster alternatives (Chapter 24) --- # Part 1: What's making it slow? ## Profiling - Use a profiler: `profvis` - A sampling/statistical profiler - Periodically stops execution and looks at the call stack --- class: hide-logo .pull-more-left[ ```r f <- function() { pause(0.1) g() h() } g <- function() { pause(0.1) h() } h <- function() { pause(0.1) } ``` ```r profvis::profvis(f()) ``` ] .pull-less-right[ 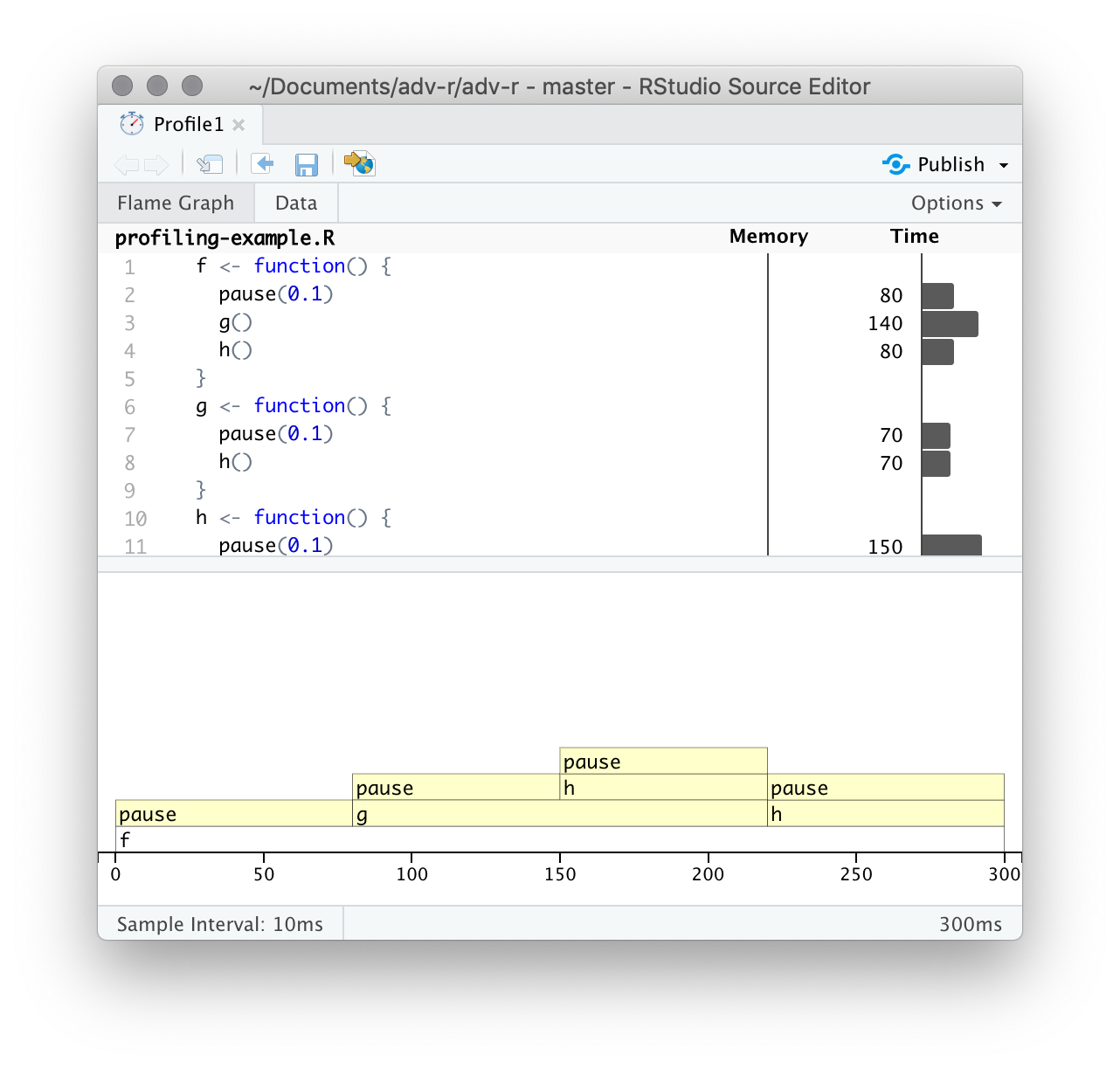 ] <hr> If you'd like to switch R version: - Run installer from CRAN - Use RSwitch utility at [http://r.research.att.com/](http://r.research.att.com/) - Bob Rudis has another version of this but I can't get it to work - Update the `R.framework/Versions/Current` directory alias directly using `ln -s` --- # Memory profiling .pull-more-left[ ```r x <- integer() for(i in 1:1e4) { x <- c(x, i) } ``` ] .pull-less-right[ 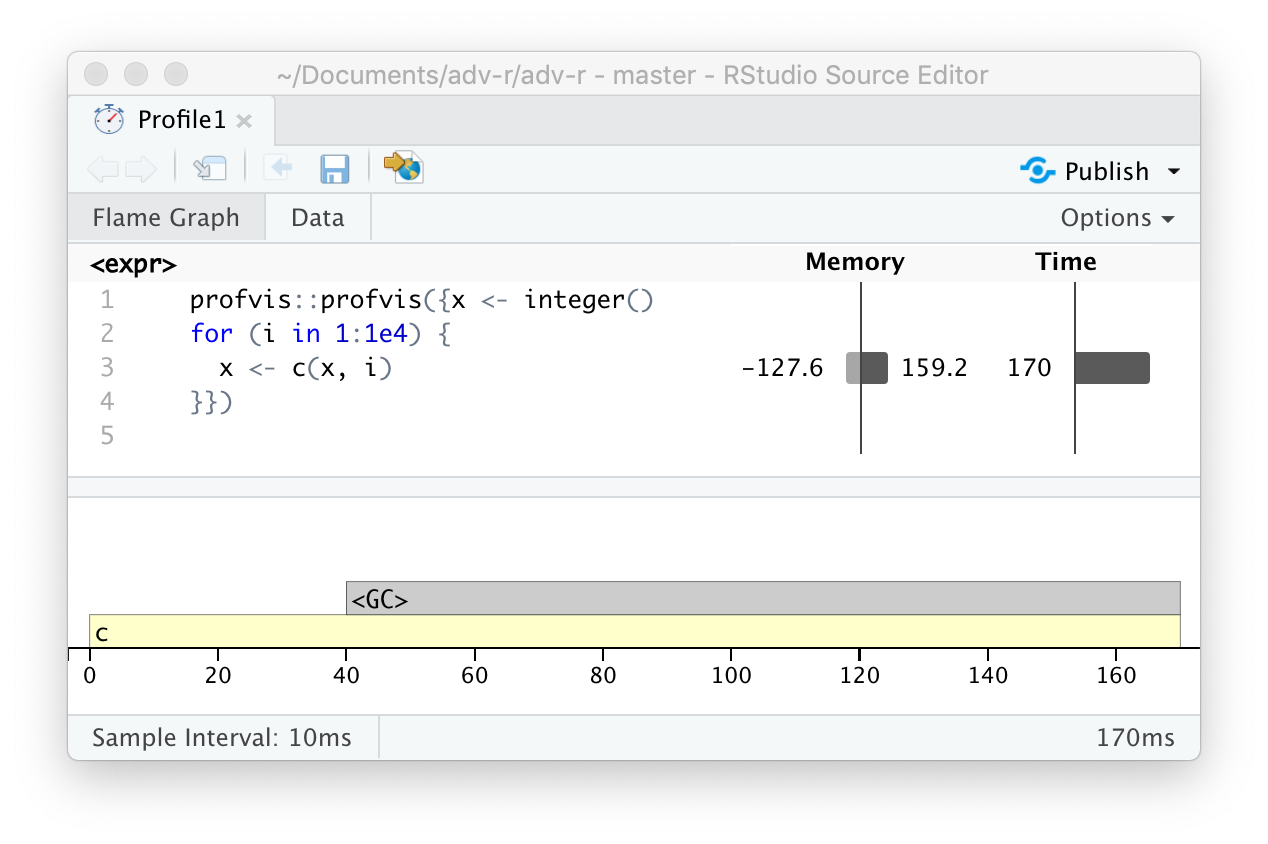 ] <hr> This shows that large amounts of memory are being allocated (bar on the right) and freed-up (bar on the left) --- # Limitations - Profiling does not extend to C code. - Using anonymous functions can make profiling difficult. Give them names. - Lazy evaluation can make things complicated: .pull-left[ ```r i <- function() { pause(0.1) 10 } j <- function(x) { x + 10 } j(i()) ``` ] .pull-right[ >"...profiling would make it seem like `i()` was called by `j()` because the argument isn't evaluated until it's needed by `j()`." ] 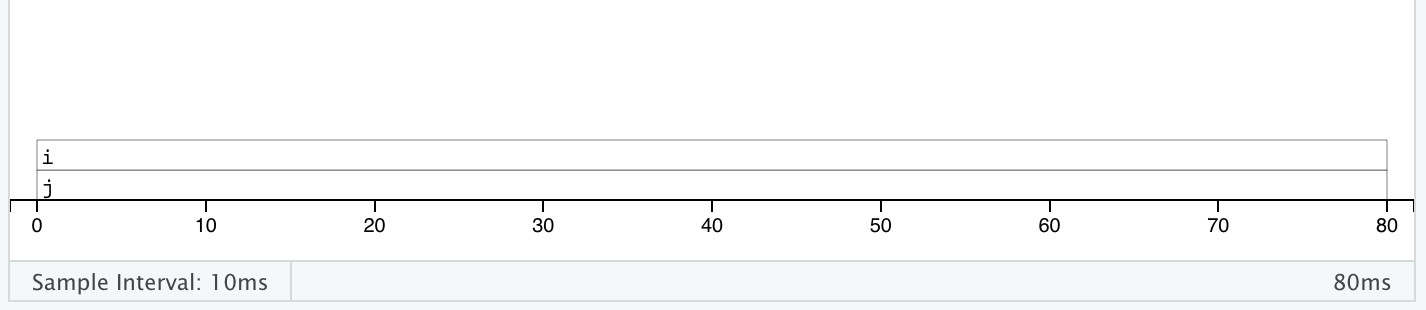 --- # Profiling Shiny apps ```r library(shiny) profvis({ runExample(example = "06_tabsets", display.mode = "normal") }) ``` --- .center[ <img src="www/shiny-profile.png" width="80%" /> ] --- # Part 1: What's making it slow? ## Microbenchmarking - For very small bits of code - **Beware of generalising to real code:** higher-order effects may mask the small bits of code > "a deep understanding of subatomic physics is not very helpful when baking" - We will use the `bench` package --- .pull-more-left[ ```r x <- runif(100) (lb <- bench::mark( sqrt(x), x ^ 0.5 )) ``` ``` ## # A tibble: 2 x 6 ## expression min median `itr/sec` mem_alloc `gc/sec` ## <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl> ## 1 sqrt(x) 334ns 700ns 232862. 848B 0 ## 2 x^0.5 2.1µs 2.25µs 269744. 848B 0 ``` ] .footnote[ median is probably the best metric to use ] --- ```r plot(lb) ``` 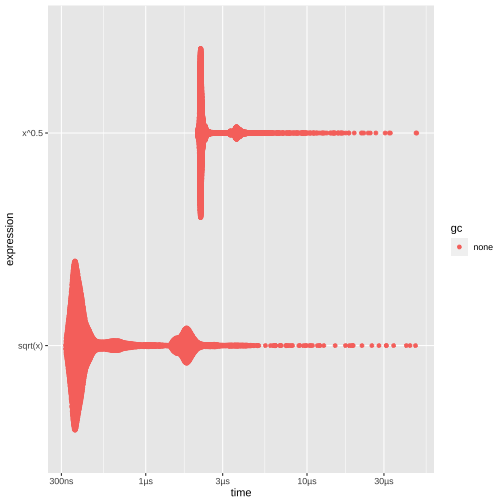<!-- --> --- # Part 2: Making it go fast! "Four" techniques: 1. Organise your code 1. Look for existing solutions 1. The importance of being lazy 1. Vectorise 1. Avoid the perils of copying data --- ## 1. Organise your code Write a function for each approach: ```r mean1 <- function(x) mean(x) mean2 <- function(x) sum(x)/length(x) ``` Generate representative test cases: ```r x <- runif(1e5) ``` Precisely compare the variants (and include unit tests (not included)) ```r bench::mark( mean1(x), mean2(x) )[c("expression", "min", "median", "itr/sec", "n_gc")] ``` ``` ## # A tibble: 2 x 4 ## expression min median `itr/sec` ## <bch:expr> <bch:tm> <bch:tm> <dbl> ## 1 mean1(x) 153.9µs 155.6µs 6133. ## 2 mean2(x) 76.2µs 76.9µs 12133. ``` --- ## 2. Check for existing solutions - CRAN task views (http://cran.rstudio.com/web/views/) - Reverse dependencies of Rcpp (https://cran.r-project.org/web/packages/Rcpp/) - Go out and talk to people: - rseek on Google (http://www.rseek.org/) - Stackoverflow with the R tag, `[R]` - https://community.rstudio.com/ - R4DS learning community!!! --- ## 3. Do as little as possible - Use a function tailered to a more specific type of input or to a more specific problem: - `rowSums()`, `colSums()`, `rowMeans()`, and `colMeans()` are faster than equivalent invocations that use `apply()` because they are vectorised - `vapply()` is faster than `sapply()` because it pre-specifies the output type - `any(x == 10)` is much faster than `10 %in% x` because testing equality is simpler than testing set inclusion. - Avoid situations where input data has to be coerced into a different type. - Example: giving a data frame to a function that requires a matrix, like `apply()` - Some other tips: - `read.csv()`: specify known column types or use `readr::read_csv()` or `data.table::fread()` - `factor()`: specify known levels - `cut()`: use `labels = FALSE` or `findInterval()` - `unlist(x, use.names = FALSE)` is faster than `unlist(x)` - `interaction()`: use `drop = TRUE` if you can --- ### Example: avoiding method dispatch ```r *x <- runif(1e2) bench::mark( mean(x), mean.default(x) )[c("expression", "min", "median", "itr/sec", "n_gc")] ``` ``` ## # A tibble: 2 x 4 ## expression min median `itr/sec` ## <bch:expr> <bch:tm> <bch:tm> <dbl> ## 1 mean(x) 2.39µs 3.5µs 251432. ## 2 mean.default(x) 1.2µs 1.39µs 533535. ``` ```r *x <- runif(1e4) bench::mark( mean(x), mean.default(x) )[c("expression", "min", "median", "itr/sec", "n_gc")] ``` ``` ## # A tibble: 2 x 4 ## expression min median `itr/sec` ## <bch:expr> <bch:tm> <bch:tm> <dbl> ## 1 mean(x) 17.6µs 18.8µs 50802. ## 2 mean.default(x) 16.3µs 16.6µs 56833. ``` --- ### But beware! <img src="www/Internal-warning.png" width="1840" /> --- ### Example 2: avoiding input coercion `as.data.frame()` is slow because it coerces each element into a data frame. You could, instead, store you data in a named list of equal-length vectors: ```r quickdf <- function(l) { class(l) <- "data.frame" attr(l, "row.names") <- .set_row_names(length(l[[1]])) l } l <- lapply(1:26, function(i) runif(1e3)) names(l) <- letters dplyr::glimpse(l[1:6]) ``` ``` ## List of 6 ## $ a: num [1:1000] 0.3726 0.9029 0.8664 0.0337 0.8816 ... ## $ b: num [1:1000] 0.946 0.109 0.767 0.237 0.614 ... ## $ c: num [1:1000] 0.659 0.938 0.317 0.414 0.152 ... ## $ d: num [1:1000] 0.559 0.888 0.872 0.917 0.669 ... ## $ e: num [1:1000] 0.933 0.923 0.757 0.407 0.272 ... ## $ f: num [1:1000] 0.452 0.533 0.915 0.198 0.259 ... ``` --- ```r bench::mark( as.data.frame = as.data.frame(l), quick_df = quickdf(l) )[c("expression", "min", "median", "itr/sec", "n_gc")] ``` ``` ## # A tibble: 2 x 4 ## expression min median `itr/sec` ## <bch:expr> <bch:tm> <bch:tm> <dbl> ## 1 as.data.frame 961.29µs 1.1ms 861. ## 2 quick_df 7.07µs 7.78µs 106802. ``` <hr> ### Caveat: This approach requires carefully reading through source code! --- ## 4. Vectorise - Finding the existing R function that is implemented in C and most closely applies to your problem - Some commonly used functions: - `rowSums()`, `colSums()`, `rowMeans()`, and `colMeans()` - Vectorised subsetting (Chapter 4) - Use `cut()` and `findInterval()` for converting continuous variables to categorical - Be aware of vectorised functions like `cumsum()` and `diff()` - Use matrix algebra - https://www.noamross.net/archives/2014-04-16-vectorization-in-r-why/ --- ## 5. Avoid copying - Often shows up if using `c()`, `append()`, `cbind()`, `rbind()`, `paste()` ```r random_string <- function() { paste(sample(letters, 50, replace = TRUE), collapse = "") } strings10 <- replicate(10, random_string()) strings100 <- replicate(100, random_string()) collapse <- function(xs) { out <- "" for (x in xs) { out <- paste0(out, x) } out } bench::mark( loop10 = collapse(strings10), loop100 = collapse(strings100), vec10 = paste(strings10, collapse = ""), vec100 = paste(strings100, collapse = ""), check = FALSE )[c("expression", "min", "median", "itr/sec", "n_gc")] ``` --- ```r bench::mark( loop10 = collapse(strings10), loop100 = collapse(strings100), vec10 = paste(strings10, collapse = ""), vec100 = paste(strings100, collapse = ""), check = FALSE )[c("expression", "min", "median", "itr/sec", "n_gc")] ``` ``` ## # A tibble: 4 x 4 ## expression min median `itr/sec` ## <bch:expr> <bch:tm> <bch:tm> <dbl> ## 1 loop10 29.01µs 30.66µs 29660. ## 2 loop100 612.86µs 644.28µs 1488. ## 3 vec10 5.37µs 5.71µs 162555. ## 4 vec100 28.17µs 29.06µs 32390. ``` --- # Case study: t-test ```r m <- 1000 n <- 50 X <- matrix(rnorm(m * n, mean = 10, sd = 3), nrow = m) grp <- rep(1:2, each = n/2) ``` --- # Case study: t-test (cont'd) Formula interface: ```r system.time( for(i in 1:m) { t.test(X[i, ] ~ grp)$statistic } ) ``` ``` ## user system elapsed ## 0.601 0.014 0.640 ``` Provide two vectors ```r system.time( for(i in 1:m) { t.test(X[i, grp == 1], X[i, grp == 2])$statistic } ) ``` ``` ## user system elapsed ## 0.140 0.002 0.143 ``` --- # Case study: t-test (cont'd) ### Add functionality to save the values: ```r compT <- function(i) { t.test(X[i, grp == 1], X[i, grp == 2])$statistic } system.time(t1 <- purrr::map_dbl(1:m, compT)) ``` ``` ## user system elapsed ## 0.140 0.002 0.143 ``` --- # Case study: t-test (cont'd) ### Do less work: ```r my_t <- function(x, grp) { t_stat <- function(x) { m <- mean(x) n <- length(x) var <- sum((x - m) ^ 2)/(n-1) list(m = m, n = n, var = var) } g1 <- t_stat(x[grp == 1]) g2 <- t_stat(x[grp == 2]) se_total <- sqrt(g1$var / g1$n + g2$var / g2$n) (g1$m - g2$m) / se_total } system.time(t2 <- purrr::map_dbl(1:m, ~ my_t(X[.,], grp))) ``` ``` ## user system elapsed ## 0.038 0.015 0.053 ``` ```r stopifnot(all.equal(t1, t2)) ``` --- # Case study: t-test (cont'd) ### Vectorise it: ```r rowtstat <- function(X, grp) { t_stat <- function(X) { m <- rowMeans(X) n <- ncol(X) var <- rowSums((X - m) ^ 2)/(n - 1) list(m = m, n = n, var = var) } g1 <- t_stat(X[, grp == 1]) g2 <- t_stat(X[, grp == 2]) se_total <- sqrt(g1$var/g1$n + g2$var/g2$n) (g1$m - g2$m) / se_total } system.time(t3 <- rowtstat(X, grp)) ``` ``` ## user system elapsed ## 7.515 0.893 0.014 ``` ```r stopifnot(all.equal(t1, t3)) ``` --- --- # Resources - [https://github.com/r-prof/jointprof](https://github.com/r-prof/jointprof) (for profiling C code) - *Evaluating the Design of the R Language* - Morandat et al., 2012 - The R Inferno (http://www.burns-stat.com/pages/Tutor/R_inferno.pdf) - Patrick Burns --- class: inverse, hide-logo # Another Slide This slide doesn't have a logo