9 Functionals
9.1 Introduction
To become significantly more reliable, code must become more transparent. In particular, nested conditions and loops must be viewed with great suspicion. Complicated control flows confuse programmers. Messy code often hides bugs. — Bjarne Stroustrup
Functionals are functions that take function as input and return a vector as output. Functionals that you probably have used before are: apply(), lapply() or tapply(). They are commonly used to replace for loops.
9.2 My first functional: map()
map() has two arguments, a vector and a function. It performs the function on each element of the vector and returns a list.
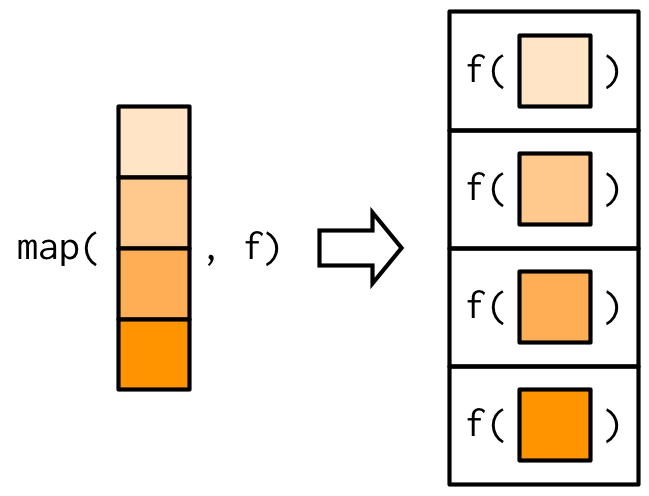
Theoretically map() just allocates a list with the necessary length and fill it with a for loop.
simple_map <- function(x, f, ...) {
out <- vector("list", length(x))
for (i in seq_along(x)) {
out[[i]] <- f(x[[i]], ...)
}
out
}Note: The base R equivalent to map() is lapply()
9.2.1 Producing atomic vectors
Sometimes it is not really necessary to have a list as an output, so you can use map_lgl(), map_int(), map_dbl(), and map_chr() to return vectors of a specified type.
9.2.2 Anonymous functions and shortcuts
map_dbl(mtcars, function(x) length(unique(x)))## mpg cyl disp hp drat wt qsec vs am gear carb
## 25 3 27 22 22 29 30 2 2 3 6Instead of typing function(x) for anonymous functions, you can use a shortcut:
map_dbl(mtcars, ~ length(unique(.x)))## mpg cyl disp hp drat wt qsec vs am gear carb
## 25 3 27 22 22 29 30 2 2 3 6In this case ~ is pronounced “twiddle”
In the following example the argument to runif() is always 2, but with map() the function is executed 3 times and the output captured in a list.
x <- map(1:3, ~ runif(2))
str(x)## List of 3
## $ : num [1:2] 0.138 0.872
## $ : num [1:2] 0.592 0.166
## $ : num [1:2] 0.927 0.671You can also use the map function to select elements (useful for working with deeply nested lists):
x <- list(
list(-1, x = 1, y = c(2), z = "a"),
list(-2, x = 4, y = c(5, 6), z = "b"),
list(-3, x = 8, y = c(9, 10, 11))
)x <- list(
list(-1, x = 1, y = c(2), z = "a"),
list(-2, x = 4, y = c(5, 6), z = "b"),
list(-3, x = 8, y = c(9, 10, 11))
)# Select by name all elements called "x"
map_dbl()map_dbl(x, "x")# Or by position all elements in the 1st position
map_dbl()map_dbl(x, 1)# Or by both the first element in "y"
map_dbl()map_dbl(x, list("y", 1))# You'll get an error if a component doesn't exist:
map_chr(x, "z")## Error: Result 1 must be a single string, not NULL of length 0# Unless you supply a .default value
map_chr(x, "z", .default = NA)## [1] NA NA NA9.2.3 Passing arguments with ...
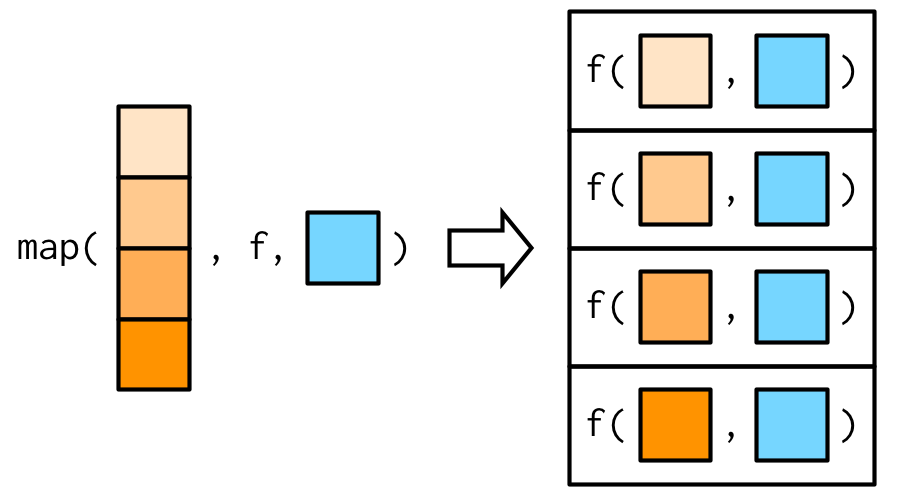
You can either add additional arguments by creating a function within map()
plus <- function(x, y) x + y
x <- c(0, 0, 0, 0)
map_dbl(x, ~ plus(.x, runif(1)))## [1] 0.8762434 0.9535156 0.3793457 0.5504553Or call the additional arguments from within map() with ...
map_dbl(x, plus, runif(1))## [1] 0.26722 0.26722 0.26722 0.26722The fine difference is, that when passing them as ... they will only be evaluated once, and not every time the funciton is called.
9.2.4 Argument names
To avoid mixing up of the function arguments of map() and the function that is called with map() the arguments of map() look a bit odd (.x and .f instead of x and f).
Hadley recommends when using map(), to use the argument names of the called function.
9.2.5 Varying another argument
- no direct way
- anonymous function:
trims <- c(0, 0.1, 0.2, 0.5)
x <- rcauchy(1000)
map_dbl(trims, function(trim) mean(x, trim = trim))## [1] 3.09524941 -0.02389633 -0.03050230 -0.069273699.2.6 Exercises
- Use
as_mapper()to explore how purrr generates anonymous functions for the integer, character, and list helpers. What helper allows you to extract attributes? Read the documentation to find out.
as_mapper(1)as_mapper(c("a", "b", "c"))as_mapper(list(1, "b", 3))as_mapper(list(1, attr_getter("a")))## function (x, ...)
## pluck(x, 1, function (x)
## attr(x, attr, exact = TRUE), .default = NULL)
## <environment: 0x7fa7ac25b200>pluck(mtcars, attr_getter("class"))## [1] "data.frame"map(1:3, ~ runif(2))is a useful pattern for generating random numbers, butmap(1:3, runif(2))is not. Why not? Can you explain why it returns the result that it does?
map(1:3, ~ runif(2))
as_mapper(~runif(2))
map(1:3, runif(2))
as_mapper(runif(2))- Use the appropriate
map()function to:
- Compute the standard deviation of every column in a numeric data frame.
df <- data.frame(a=runif(10), b=1:10, c=c(rep(100, 10)))map(df, sd)- Compute the standard deviation of every numeric column in a mixed data frame. (Hint: you’ll need to do it in two steps.)
df <- data.frame(num1=runif(10), num2=1:10, char1=letters[1:10])df[map_lgl(df,is.numeric)] %>% map(sd)- Compute the number of levels for every factor in a data frame.
df <- data.frame(a=as.factor(sample(letters, size = 12, replace=TRUE)), b=as.factor(sample(letters, size = 12, replace=TRUE)), c=as.factor(sample(letters, size = 12, replace=TRUE)))map(df, levels) %>% map(length)- The following code simulates the performance of a t-test for non-normal data. Extract the p-value from each test, then visualise.
trials <- map(1:100, ~ t.test(rpois(10, 10), rpois(7, 10)))map_dbl(trials, "p.value") %>% hist()- The following code uses a map nested inside another map to apply a function to every element of a nested list. Why does it fail, and what do you need to do to make it work?
x <- list(
list(1, c(3, 9)),
list(c(3, 6), 7, c(4, 7, 6))
)
triple <- function(x) x * 3
map(x, map, .f = triple)map(x, .f = map, triple)The second map here would be one of the ... arguments of the first map, so passed on as an argument to .f (triple).
- Use
map()to fit linear models to themtcarsdataset using the formulas stored in this list:
formulas <- list(
mpg ~ disp,
mpg ~ I(1 / disp),
mpg ~ disp + wt,
mpg ~ I(1 / disp) + wt
)map(formulas, lm, data=mtcars)- Fit the model
mpg ~ dispto each of the bootstrap replicates ofmtcarsin the list below, then extract the \(R^2\) of the model fit (Hint: you can compute the \(R^2\) withsummary().)
bootstrap <- function(df) {
df[sample(nrow(df), replace = TRUE), , drop = FALSE]
}
bootstraps <- map(1:10, ~ bootstrap(mtcars))map(bootstraps, ~lm(mpg~disp, data=.x))9.4 Map variants
In total there are 23 map() variants.
- Output same type as input with
modify() - Iterate over two inputs with
map2() - Iterate with an index using
imap() - Return nothing with
walk() - Iterate over any number of inputs with
pmap()
| List | Atomic | Same type | Nothing | |
|---|---|---|---|---|
| One argument | map() | map_lgl(), … | modify() | walk() |
| Two arguments | map2() | map2_lgl(), … | modify2() | walk2() |
| One argument + index | imap() | imap_lgl(), … | imodify() | iwalk() |
| N arguments | pmap() | pmap_lgl(), … | — | pwalk() |
9.4.1 Same type of output as input: modify()
df <- data.frame(
x = 1:3,
y = 6:4
)With map():
map(df, ~ .x * 2)## $x
## [1] 2 4 6
##
## $y
## [1] 12 10 8… returns a list.
modify()
modify(df, ~ .x * 2)… returns the same type of output as input.
9.4.2 Two inputs: map2() and friends
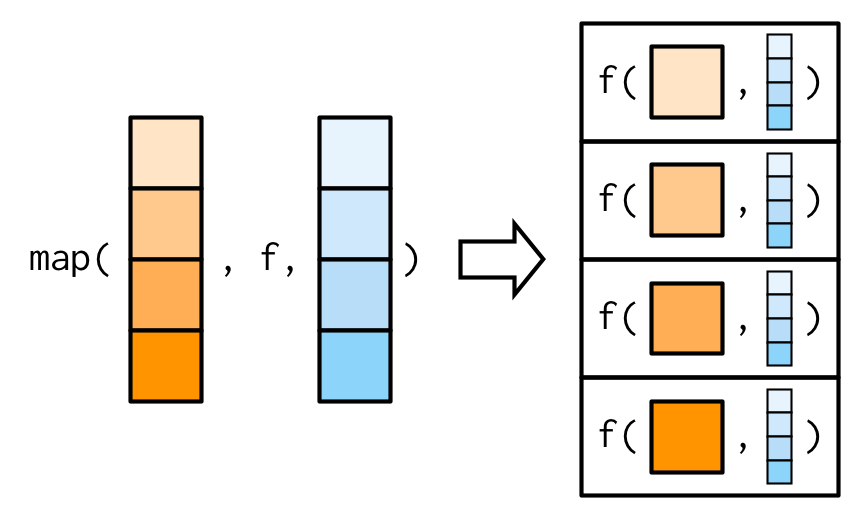
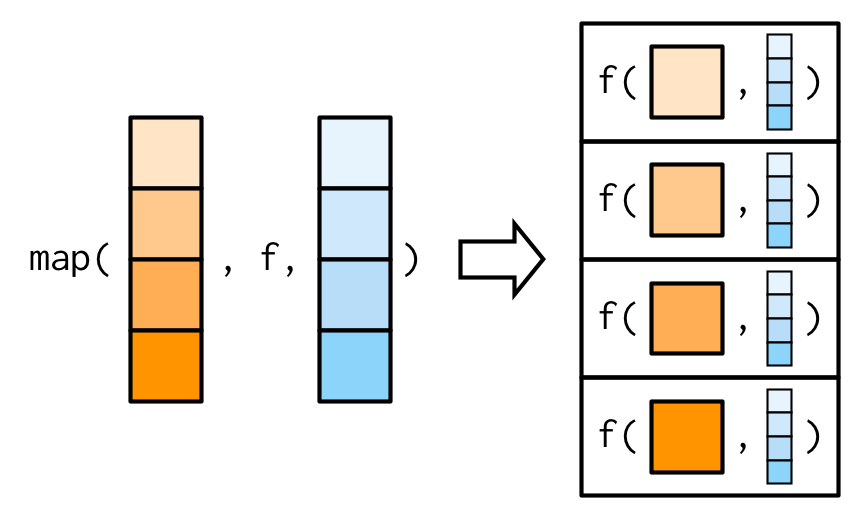
The main difference between map() and map2()is that map2() is vectorised over two arguments:
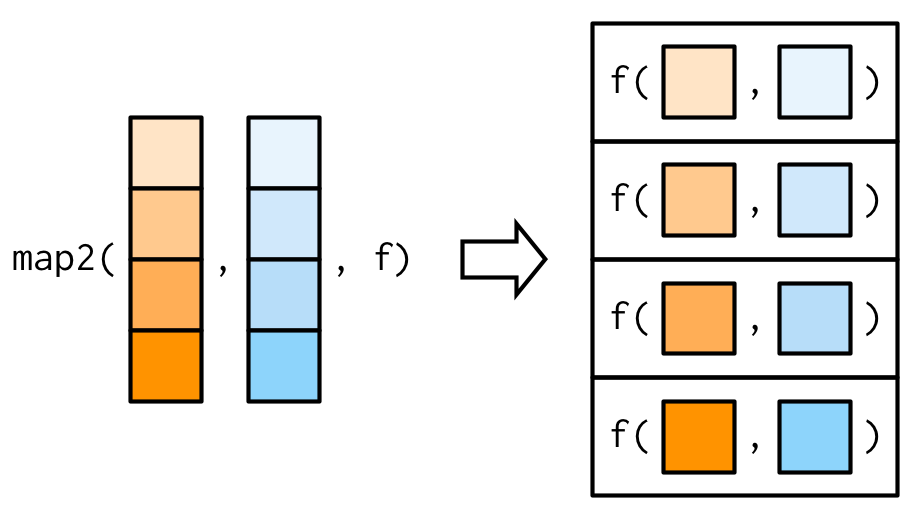
9.4.3 No outputs: walk() and friends
Mainly for functions that are called because of their side-effects
welcome <- function(x) {
cat("Welcome ", x, "!\n", sep = "")
}
names <- c("Hadley", "Jenny")map() also returns the NULL values:
map(names, welcome)## Welcome Hadley!
## Welcome Jenny!## [[1]]
## NULL
##
## [[2]]
## NULLwalk() functions ignore the value of the function:
walk(names, welcome)## Welcome Hadley!
## Welcome Jenny!9.4.4 Iterating over values and indices
The imap() family let’s you iterate over indices or names.
legs <- c(8,6,4,2,1)
names(legs) <- c("Spider", "Ant", "Cat", "Human", "Pirate")imap_chr(legs, ~ paste0("A ", .y, " has ", .x, " legs"))## Spider Ant Cat
## "A Spider has 8 legs" "A Ant has 6 legs" "A Cat has 4 legs"
## Human Pirate
## "A Human has 2 legs" "A Pirate has 1 legs"legs <- unname(legs)
imap_chr(legs, ~paste0("Nr. ", .y, " has ", .x, " legs"))## [1] "Nr. 1 has 8 legs" "Nr. 2 has 6 legs" "Nr. 3 has 4 legs" "Nr. 4 has 2 legs"
## [5] "Nr. 5 has 1 legs"Useful for:
- constructing labels
- work with the values along with their positions
9.4.5 Any number of inputs: pmap() and friends
With the pmap() family functions you can iterate over n vectorised inputs that are stored in a list (best: named):
params <- tibble::tribble(
~ n, ~ min, ~ max,
1L, 0, 1,
2L, 10, 100,
3L, 100, 1000
)
pmap(params, runif)## [[1]]
## [1] 0.1805052
##
## [[2]]
## [1] 43.60020 80.45185
##
## [[3]]
## [1] 101.9223 493.8894 866.70479.4.6 Exercises
- Explain the results of
modify(mtcars, 1).
head(modify(mtcars, 1))
map(mtcars, 1)Modify has to return the same structure as .x, in this case a data frame with dimensions 32, 11. In this case the first row is plucked from the data frame (as with map()) and returned in the same format as the input
- Rewrite the following code to use
iwalk()instead ofwalk2(). What are the advantages and disadvantages?
temp <- tempfile()
dir.create(temp)
cyls <- split(mtcars, mtcars$cyl)
paths <- file.path(temp, paste0("cyl-", names(cyls), ".csv"))
walk2(cyls, paths, write.csv)
dir(temp)names(cyls) <- file.path(temp, paste0("cyl-", names(cyls), ".csv"))
iwalk(cyls, write.csv)
dir(temp)- Explain how the following code transforms a data frame using functions stored in a list.
trans <- list(
disp = function(x) x * 0.0163871,
am = function(x) factor(x, labels = c("auto", "manual"))
)
nm <- names(trans)
mtcars[nm] <- map2(trans, mtcars[nm], function(f, var) f(var))- Compare and contrast the
map2()approach to thismap()approach:
mtcars[vars] <- map(vars, ~ trans[[.x]](mtcars[[.x]]))- What does
write.csv()return? i.e. what happens if you use it withmap2()instead ofwalk2()?
9.5 Reduce family
9.5.1 Basics
genes <- c("MKI67", "SOX2", "HES1", "HES5", "NES", "PAX6", "TUBB3", "STMN1", "FAT3", "DCX")
l <- map(1:4, ~ sample(genes, 15, replace = T))out <- l[[1]]
out <- intersect(out, l[[2]])
out <- intersect(out, l[[3]])
out <- intersect(out, l[[4]])
out## [1] "FAT3" "PAX6" "DCX" "HES1"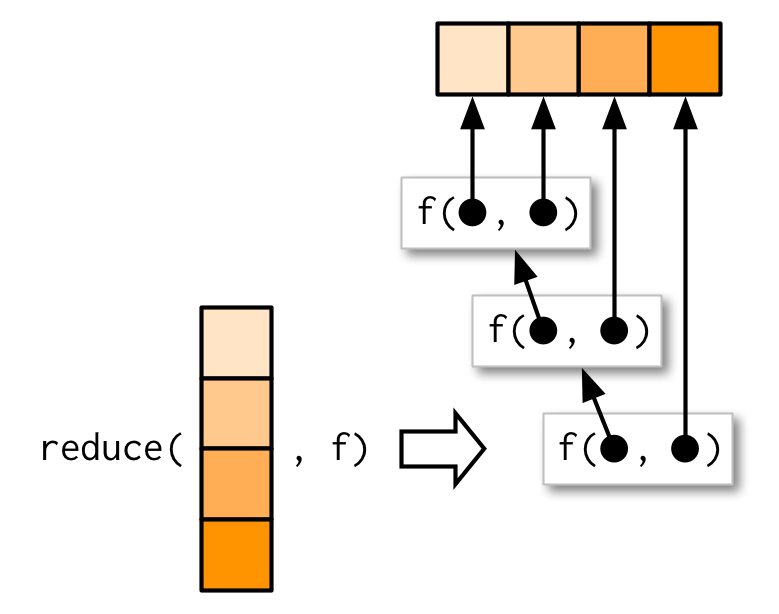
reduce(l, intersect)## [1] "FAT3" "PAX6" "DCX" "HES1"reduce(l, union)## [1] "FAT3" "STMN1" "SOX2" "HES5" "PAX6" "DCX" "HES1" "NES" "MKI67"
## [10] "TUBB3"9.5.2 Accumulate
accumulate(l, intersect)## [[1]]
## [1] "FAT3" "STMN1" "SOX2" "HES5" "SOX2" "PAX6" "DCX" "STMN1" "HES1"
## [10] "SOX2" "HES5" "SOX2" "HES1" "NES" "HES5"
##
## [[2]]
## [1] "FAT3" "SOX2" "HES5" "PAX6" "DCX" "HES1"
##
## [[3]]
## [1] "FAT3" "SOX2" "PAX6" "DCX" "HES1"
##
## [[4]]
## [1] "FAT3" "PAX6" "DCX" "HES1"9.5.3 Output types
If the x provided is of length 1 or zero the reduce function either returns the input value or asks for an .init value
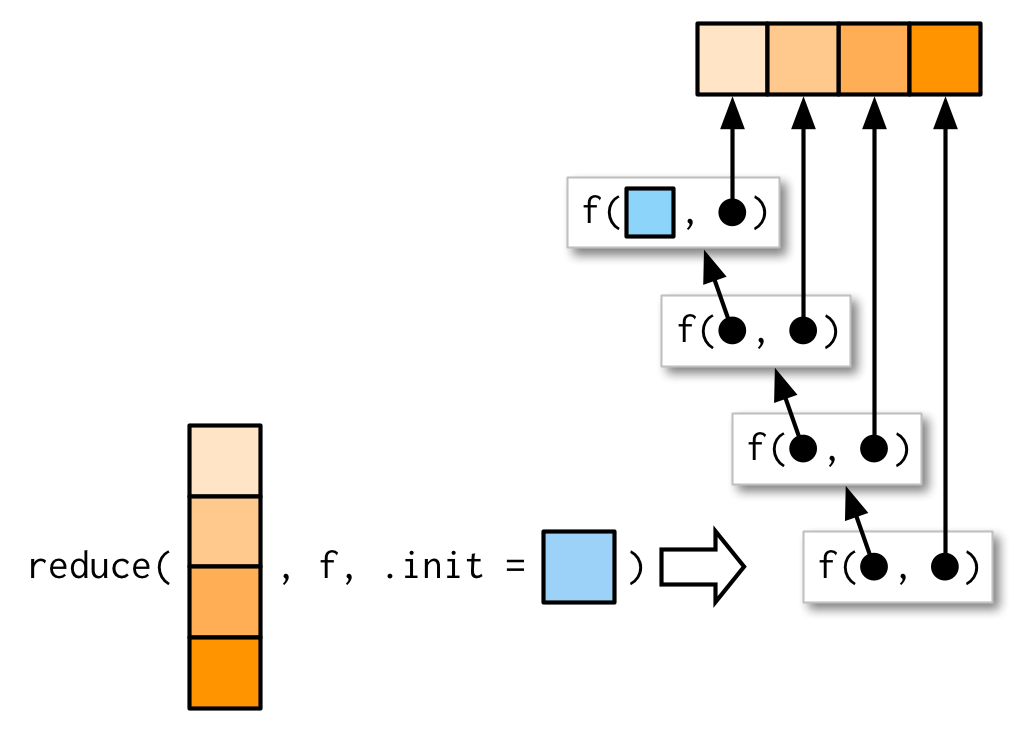
If you’re using reduce() in a function, you should always supply .init. Think carefully about what your function should return when you pass a vector of length 0 or 1, and make sure to test your implementation.
9.6 Predicate functionals
A predicate is a function that returns a single
TRUEorFALSE, likeis.character(),is.null(), orall(), and we say a predicate matches a vector if it returnsTRUE.
9.6.1 Basics
purrr has six predicate functions:
df <- data.frame(a=c(1.2, 2.3, 4.5), b=c(8,3.4,2.6))
trans <- list(
disp = function(x) x * 0.0163871,
am = function(x) factor(x, labels = c("auto", "manual"))
)
nm <- names(trans)
mtcars[nm] <- map2(trans, mtcars[nm], function(f, var) f(var))some(mtcars, is.double)
some(mtcars, is.logical)every(mtcars, is.double)
every(df, is.double)detect(mtcars, is.factor)
detect_index(mtcars, is.factor)keep(mtcars, is.factor)
discard(mtcars, is.double)9.6.2 Map variants
df <- data.frame(
num1 = c(0, 10, 20),
num2 = c(5, 6, 7),
chr1 = c("a", "b", "c"),
stringsAsFactors = FALSE
)
str(map_if(df, is.numeric, mean))
str(modify_if(df, is.numeric, mean))
str(map(keep(df, is.numeric), mean))9.6.3 Exercises
- Why isn’t
is.na()a predicate function? What base R function is closest to being a predicate version ofis.na()?
my_vector <- c(0, 10, 20, NA)
is.na(my_vector)
is.na(my_vector)any(is.na(my_vector))simple_reduce()has a problem whenxis length 0 or length 1. Describe the source of the problem and how you might go about fixing it.
simple_reduce <- function(x, f) {
out <- x[[1]]
for (i in seq(2, length(x))) {
out <- f(out, x[[i]])
}
out
}simple_reduce(1, `+`)- Implement the
span()function from Haskell: given a listxand a predicate functionf,span(x, f)returns the location of the longest sequential run of elements where the predicate is true. (Hint: you might findrle()helpful.)
# Just some example df to work with
df <- data.frame(
num1 = c(0, 10, 20),
num2 = c(5, 6, 7),
chr1 = c("a", "b", "c"),
stringsAsFactors = FALSE
)
df2 <- sample(df, 20, replace=TRUE)span <- function(x,f){
out <- map_lgl(x, f) %>% as.numeric() %>% rle()
out.true <- out$lengths==max(out$length[which(out$values==1)])
a <- 0
out.span <- for(i in seq_along(out.true))
if(!out.true[i]){
a <- a+ out$lengths[i]
}else{
a <- a+1
b <- a + out$lengths[i]
break
}
out.span <- a:b
out.span
}
span(df2, is.double)- Implement
arg_max(). It should take a function and a vector of inputs, and return the elements of the input where the function returns the highest value. For example,arg_max(-10:5, function(x) x ^ 2)should return -10.arg_max(-5:5, function(x) x ^ 2)should returnc(-5, 5). Also implement the matchingarg_min()function.
x <- -10:5
y <- -5:5
arg_max <- function(x, f){
out <- f(x)
out <- which(out==max(out))
x[out]
}
arg_max(x, function(x) x^2)
arg_max(y, function(x) x^2)
arg_min <- function(x, f){
out <- f(x)
out <- which(out==min(out))
x[out]
}
arg_min(x, function(x) x/2)
arg_min(y, function(x) x/2)- The function below scales a vector so it falls in the range [0, 1]. How would you apply it to every column of a data frame? How would you apply it to every numeric column in a data frame?
scale01 <- function(x) {
rng <- range(x, na.rm = TRUE)
(x - rng[1]) / (rng[2] - rng[1])
}# modify(df, scale01)
modify_if(df, is.numeric, scale01)9.7 Base functionals
- functionals without equivalent in the
purrrpackage - more useful in mathematica/statistics
- not so useful in data analysis